
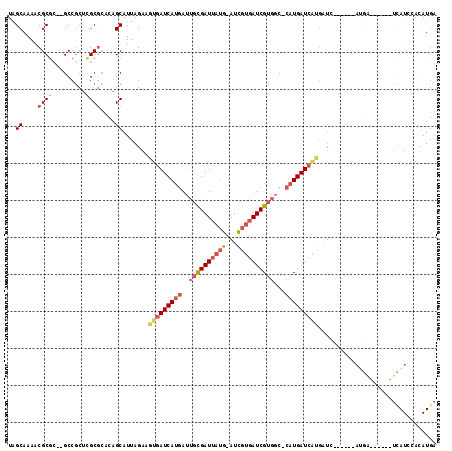
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,356,693 – 11,356,831 |
| Length | 138 |
| Max. P | 0.999375 |

| Location | 11,356,693 – 11,356,805 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.35 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11356693 112 + 22224390 UGGCAAAACGCGC--GCCGCUCGCGCACAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG-AUCGUGAUCGUGGC-CAUGAUCAUGAUUAUGAUUAUGAUCAUGAUCAUCCACAUGA (((......((((--(.....)))))............((((((((((((..(((((((-(((((((((((...-.))))))))))))))))))..)))))))))))))))..... ( -48.80) >DroSec_CAF1 17645 106 + 1 UAGCAAAACGCGC--GCCGCUCGCGCACAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG-AUCGUGAUCGUGGC-CAUGAUCAUGAUU------AUGAUCAUGAUCAUCCACAUGA .........((((--(.....)))))....(((.....((((((((((((..(((((((-((((((........-)))))))))))))------..))))))))))))....))). ( -41.60) >DroSim_CAF1 239 94 + 1 UAGCAAAACGCGC--GCCGCUCGCGCACAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG-AUCGUGAUCGUGGC-CAUGAUC------------------AUGAUCAUCCACAUGA .........((((--(.....))))).........((.(((((((((.(..(((((((.-...)))))))..).-)))))))------------------))..)).......... ( -33.00) >DroEre_CAF1 21333 100 + 1 UAGCAAAACGCAC--GCCGCUCGCGCGCAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG-AUCGUGAUCGUGGC-CAUGAUCAUGAUC------AUGA------UUAUGAACAUGA .........((.(--((.....))).))..............(((((.((..(((((((-(((((((((((...-.))))))))))))------))))------))..)).))))) ( -36.40) >DroYak_CAF1 22608 100 + 1 UAGCAAAACGCGC--GCCGCUUGCGCACAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG-AUCGUGAUCGUGGC-CAUGAUCAUGAUC------AUGA------UCAUGAACAUGA ..((.....))((--((.....))))................(((((.((..(((((((-(((((((((((...-.))))))))))))------))))------))..)).))))) ( -38.60) >DroMoj_CAF1 28089 94 + 1 UAGCAAAACGCGACAGCCAAAUGC-----GCAUUAGAAGUCAUCAAAAACAUGAUUG-GAAUUGGUAUCG--GCAAAUGAUAGCGGUA------AAAU------AUAUCGAUAU-- ..((.....))....(((..((((-----.((.....((((((.......)))))).-....)))))).)--)).........(((((------....------.)))))....-- ( -15.20) >consensus UAGCAAAACGCGC__GCCGCUCGCGCACAGCAUUAGAAGUGAUCAUGAUUGCGAUUAUG_AUCGUGAUCGUGGC_CAUGAUCAUGAUC______AUGA______UCAUCCACAUGA ..((....((((.........))))....)).......(((((((((..((((((((((...))))))))))...)))))))))................................ (-19.01 = -20.35 + 1.34)


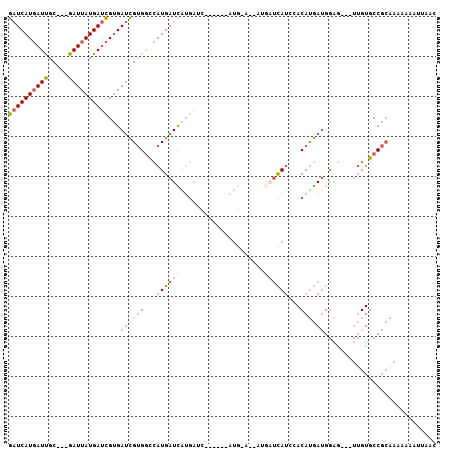
| Location | 11,356,731 – 11,356,831 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.13 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11356731 100 + 22224390 GAUCAUGAUUGC---GAUUAUGAUCGUGAUCGUGGCCAUGAUCAUGAUUAUGAUUAUG-AUCAUGAUCAUCCACAUGAUGGAG---UUGUGCCGCAAAAAAAUUAAC ((((((((((..---......))))))))))((((((((((((((((((((....)))-))))))))))((((.....)))).---.)).)))))............ ( -40.40) >DroSec_CAF1 17683 94 + 1 GAUCAUGAUUGC---GAUUAUGAUCGUGAUCGUGGCCAUGAUCAUGAUU------AUG-AUCAUGAUCAUCCACAUGAUGGAG---UUGUGCCGCAAAAAAAUUAAC ((((((((((..---......))))))))))(((((((((((((((((.------...-))))))))))((((.....)))).---.)).)))))............ ( -37.30) >DroSim_CAF1 277 82 + 1 GAUCAUGAUUGC---GAUUAUGAUCGUGAUCGUGGCCAUGAUC-------------------AUGAUCAUCCACAUGAUGGAG---UUGUGCCGCAAAAAAAUUAAC ((((((((((..---......))))))))))((((((((.(((-------------------(((........)))))).)..---.)).)))))............ ( -27.10) >DroEre_CAF1 21371 88 + 1 GAUCAUGAUUGC---GAUUAUGAUCGUGAUCGUGGCCAUGAUCAUGAUC------AUG-A------UUAUGAACAUGAUGGAG---UUGUGCCGCGAAGAAAUUAAC .((((((.((..---((((((((((((((((((....))))))))))))------)))-)------))..)).))))))((..---.....)).............. ( -31.80) >DroYak_CAF1 22646 88 + 1 GAUCAUGAUUGC---GAUUAUGAUCGUGAUCGUGGCCAUGAUCAUGAUC------AUG-A------UCAUGAACAUGAUGGAG---UUGUGCCGCGAAGAAAUUAAC .((((((.((..---((((((((((((((((((....))))))))))))------)))-)------))..)).))))))((..---.....)).............. ( -33.30) >DroAna_CAF1 12466 92 + 1 AGUCAUUAUCGCCGUGAUUAUGAAUGCGAAUGCGAUUAUGAUUUGGAAA------CUGAAUUAUAAGCA--CACACGACGAGACGAGAGUGCCCGGAAUG------- .(((....(((.((((.........((....))(.((((((((((....------).))))))))).).--..)))).))))))................------- ( -18.30) >consensus GAUCAUGAUUGC___GAUUAUGAUCGUGAUCGUGGCCAUGAUCAUGAUC______AUG_A__AUGAUCAUCCACAUGAUGGAG___UUGUGCCGCAAAAAAAUUAAC ((((((((((.....))))))))))...((((((...((((((.....................))))))...))))))............................ (-14.03 = -14.87 + 0.84)


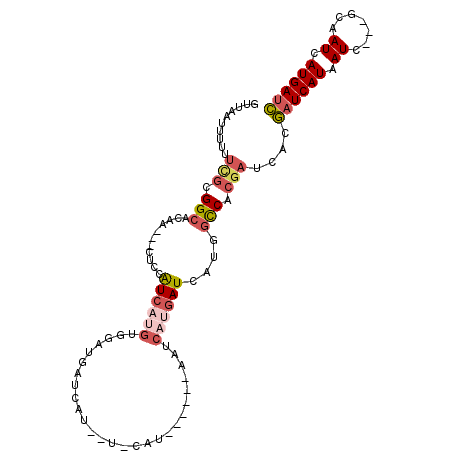
| Location | 11,356,731 – 11,356,831 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -5.08 |
| Energy contribution | -6.19 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11356731 100 - 22224390 GUUAAUUUUUUUGCGGCACAA---CUCCAUCAUGUGGAUGAUCAUGAU-CAUAAUCAUAAUCAUGAUCAUGGCCACGAUCACGAUCAUAAUC---GCAAUCAUGAUC ..........(((((((.((.---.(((((...)))))((((((((((-.((....)).)))))))))))))))..(((....)))......---))))........ ( -26.00) >DroSec_CAF1 17683 94 - 1 GUUAAUUUUUUUGCGGCACAA---CUCCAUCAUGUGGAUGAUCAUGAU-CAU------AAUCAUGAUCAUGGCCACGAUCACGAUCAUAAUC---GCAAUCAUGAUC ..........(((((((.((.---.(((((...)))))((((((((((-...------.)))))))))))))))..(((....)))......---))))........ ( -26.50) >DroSim_CAF1 277 82 - 1 GUUAAUUUUUUUGCGGCACAA---CUCCAUCAUGUGGAUGAUCAU-------------------GAUCAUGGCCACGAUCACGAUCAUAAUC---GCAAUCAUGAUC ..........((((((.....---.(((((...)))))(((((.(-------------------((((........))))).)))))...))---))))........ ( -20.70) >DroEre_CAF1 21371 88 - 1 GUUAAUUUCUUCGCGGCACAA---CUCCAUCAUGUUCAUAA------U-CAU------GAUCAUGAUCAUGGCCACGAUCACGAUCAUAAUC---GCAAUCAUGAUC ..........(((.(((.((.---....((((((.((((..------.-.))------)).))))))..))))).)))....((((((.((.---...)).)))))) ( -20.20) >DroYak_CAF1 22646 88 - 1 GUUAAUUUCUUCGCGGCACAA---CUCCAUCAUGUUCAUGA------U-CAU------GAUCAUGAUCAUGGCCACGAUCACGAUCAUAAUC---GCAAUCAUGAUC ..........(((.(((..((---(........)))(((((------(-(((------....)))))))))))).)))....((((((.((.---...)).)))))) ( -23.50) >DroAna_CAF1 12466 92 - 1 -------CAUUCCGGGCACUCUCGUCUCGUCGUGUG--UGCUUAUAAUUCAG------UUUCCAAAUCAUAAUCGCAUUCGCAUUCAUAAUCACGGCGAUAAUGACU -------......((((((.(.((......)).).)--))))).........------........((((.(((((...................))))).)))).. ( -17.61) >consensus GUUAAUUUUUUCGCGGCACAA___CUCCAUCAUGUGGAUGAUCAU__U_CAU______AAUCAUGAUCAUGGCCACGAUCACGAUCAUAAUC___GCAAUCAUGAUC ..........(((.(((...........((((((...........................))))))....))).)))....((((((.((.......)).)))))) ( -5.08 = -6.19 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:44 2006