
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,353,083 – 11,353,216 |
| Length | 133 |
| Max. P | 0.984652 |

| Location | 11,353,083 – 11,353,176 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11353083 93 - 22224390 AAAUAGUAAAAAUGCCCCAUCAUAUGUACAUACAUACAUAUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAAAAAUUUUAAUUUAUG ....((((.......(((((.(((((((......))))))))))))..(((((.....))))))))).......................... ( -19.80) >DroSec_CAF1 14049 93 - 1 AUAUAGUAAAAAUGCCCCAUCAUAUGUACAUUCAUGCAUAUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAAAAAUUUUAAUUUAUG ....((((.......(((((.(((((((......))))))))))))..(((((.....))))))))).......................... ( -19.80) >DroEre_CAF1 17644 77 - 1 AAAUAGUAAAAAUGCCCCAUCAU----------------AUAUGGGAUGUGAGAAGUUCUCGCUGCUUCAAACAAUAAAAAUUUUAAUUUAUG .....(((((.....(((((...----------------..))))).(((..(((((.......)))))..))).............))))). ( -13.30) >DroYak_CAF1 18122 77 - 1 AAAUAGUAAAAAUGGCCCAUCAU----------------AUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAAAAAUUUUAAUUUAUG ............((((((((...----------------..)))))..(((((.....)))))....)))....................... ( -13.50) >consensus AAAUAGUAAAAAUGCCCCAUCAU________________AUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAAAAAUUUUAAUUUAUG ....((((.......(((((.(((((((......))))))))))))..(((((.....))))))))).......................... (-15.16 = -15.73 + 0.56)

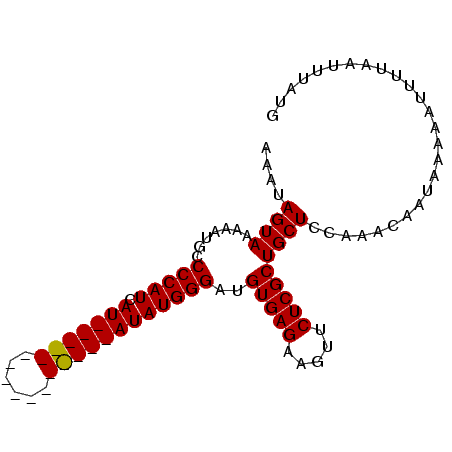

| Location | 11,353,098 – 11,353,216 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11353098 118 + 22224390 UUAUUGUUUGGAGCAGCGAGAACUUCUCACAUCCCAUAUAUGUAUGUAUGUACAUAUGAUGGGGCAUUUUUACUAUUUAAAUAUUAUUGUUAUUGCUUCUUCAUCUUUUGGUGAUUUC .........(((((((((((.....)))....(((((((((((((....)))))))).)))))))..............((((....))))..)))))).(((((....))))).... ( -30.60) >DroSec_CAF1 14064 118 + 1 UUAUUGUUUGGAGCAGCGAGAACUUCUCACAUCCCAUAUAUGCAUGAAUGUACAUAUGAUGGGGCAUUUUUACUAUAUAAAUAUUAUUGUUGUUGCUUCUUCAUCUUUCGGUGGAUUC .........(((((((((((.....)))...(((((((((((.((....)).))))).))))))...........................))))))))((((((....))))))... ( -28.70) >DroEre_CAF1 17659 102 + 1 UUAUUGUUUGAAGCAGCGAGAACUUCUCACAUCCCAUAU----------------AUGAUGGGGCAUUUUUACUAUUUAAAUAUUAUUGUUAUUGCUUCUCCAUCUUUUGGUGAUUUC .........(((((((((((.....)))....(((((..----------------...)))))))..............((((....))))..))))))..((((....))))..... ( -21.70) >DroYak_CAF1 18137 102 + 1 UUAUUGUUUGGAGCAGCGAGAACUUCUCACAUCCCAUAU----------------AUGAUGGGCCAUUUUUACUAUUUAAAUAUUAUUGUUAUUGCUUCUACAUCUUUUGAUGAUUUC .........(((((((.(((.....)))....(((((..----------------...))))).............................)))))))..((((....))))..... ( -19.80) >consensus UUAUUGUUUGGAGCAGCGAGAACUUCUCACAUCCCAUAU________________AUGAUGGGGCAUUUUUACUAUUUAAAUAUUAUUGUUAUUGCUUCUUCAUCUUUUGGUGAUUUC .........(((((((.(((.....)))....(((((((((((((....)))))))).))))).............................)))))))..((((....))))..... (-21.33 = -21.95 + 0.62)

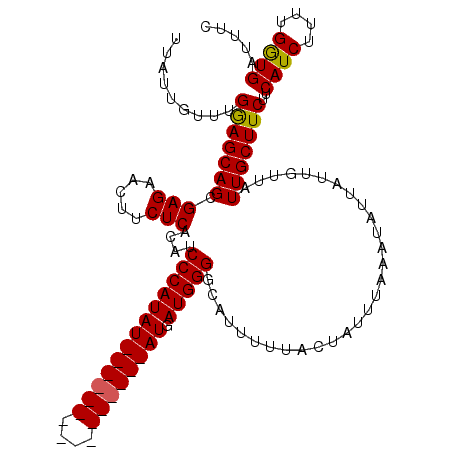
| Location | 11,353,098 – 11,353,216 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11353098 118 - 22224390 GAAAUCACCAAAAGAUGAAGAAGCAAUAACAAUAAUAUUUAAAUAGUAAAAAUGCCCCAUCAUAUGUACAUACAUACAUAUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAA ...................(.((((..............................(((((.(((((((......))))))))))))..(((((.....))))))))).)......... ( -22.80) >DroSec_CAF1 14064 118 - 1 GAAUCCACCGAAAGAUGAAGAAGCAACAACAAUAAUAUUUAUAUAGUAAAAAUGCCCCAUCAUAUGUACAUUCAUGCAUAUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAA .....((.(....).))..(.((((..............................(((((.(((((((......))))))))))))..(((((.....))))))))).)......... ( -24.80) >DroEre_CAF1 17659 102 - 1 GAAAUCACCAAAAGAUGGAGAAGCAAUAACAAUAAUAUUUAAAUAGUAAAAAUGCCCCAUCAU----------------AUAUGGGAUGUGAGAAGUUCUCGCUGCUUCAAACAAUAA .......(((.....))).((((((..............................(((((...----------------..)))))..(((((.....)))))))))))......... ( -22.00) >DroYak_CAF1 18137 102 - 1 GAAAUCAUCAAAAGAUGUAGAAGCAAUAACAAUAAUAUUUAAAUAGUAAAAAUGGCCCAUCAU----------------AUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAA .....((((....))))..(.((((..............................(((((...----------------..)))))..(((((.....))))))))).)......... ( -18.00) >consensus GAAAUCACCAAAAGAUGAAGAAGCAAUAACAAUAAUAUUUAAAUAGUAAAAAUGCCCCAUCAU________________AUAUGGGAUGUGAGAAGUUCUCGCUGCUCCAAACAAUAA ...................(.((((..............................(((((.(((((((......))))))))))))..(((((.....))))))))).)......... (-18.16 = -18.73 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:42 2006