| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,351,740 – 11,351,839 |
| Length | 99 |
| Max. P | 0.986853 |

| Location | 11,351,740 – 11,351,839 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
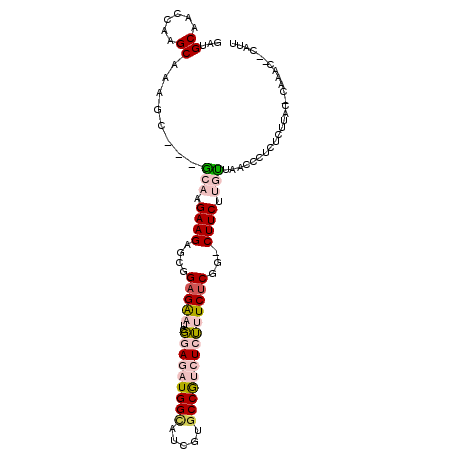
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11351740 99 + 22224390 GAUGCAACCAAGCAAAGC---GCAAGAAGAGCGGAGAAUCGGAGAUGGCAUCGUACCGUCUCUUUCUCGG-CUUCUUGUUAACCCUCUCGUACGCAAAC--CAUU ..(((......)))..((---((((((..(((.(((((..((((((((.......))))))))))))).)-))))))))..((......))..))....--.... ( -30.40) >DroPse_CAF1 13411 97 + 1 GAUGCACCCAAGCCGAACCGAGAACGAAGA--GGAGGAAGCCAGAUGGUAAAUAGCCAGAUCGCUCUCUGCCUUCUCGCUGAAUCGCUCUUGG----GG--CAUU (((((.((((((.(((..(.((..(((..(--((((..(((.(..((((.....))))..).)))..)).)))..))))))..)))..)))))----))--)))) ( -36.10) >DroSec_CAF1 12690 99 + 1 GAUGCAACCAAGCAAAGC---GCAAGAAGAGCGGAGAAUCGGAGAUGGCAUCGUGCCGUCUCUUUCUCGG-CUUCUUGUUAACCAUCUCGUACGCAAAC--CAUU ..(((......)))..((---((((((..(((.(((((..(((((((((.....)))))))))))))).)-))))))))..((......))..))....--.... ( -34.80) >DroEre_CAF1 16327 99 + 1 GAUGCAACCAAGCAAAGC---ACAAGAAGAGCGGAGAAUCGGAGAUGGCAUCAUGCCGUCUCUUUCUCGG-CUUCUUGUUAACCCUCUCUUAUCCAAAC--CAUU ..(((......)))....---((((((..(((.(((((..(((((((((.....)))))))))))))).)-))))))))....................--.... ( -33.60) >DroYak_CAF1 16798 101 + 1 GAUGCAACCAAGCAAAGC---GCAAGAAGAGCGGCGAAUCGGAGAUGGCAUCGUGCCGUCUCUUUCUCGG-CUUCCUGUUAACCAUCUCUUACCCAAACCCCAUU ((((.(((.((((....(---((.......))).(((...(((((((((.....)))))))))...))))-)))...)))...)))).................. ( -27.10) >DroPer_CAF1 13296 97 + 1 GAUGCACCCAAGCCGAACCGAGAACGAAGA--GGAGGAAGCCAGAUGGUAAAUAGCCAGAUCGCUCUCUGCCUUCUCGCUGAAUCGCUCUUGG----GG--CAUU (((((.((((((.(((..(.((..(((..(--((((..(((.(..((((.....))))..).)))..)).)))..))))))..)))..)))))----))--)))) ( -36.10) >consensus GAUGCAACCAAGCAAAGC___GCAAGAAGAGCGGAGAAUCGGAGAUGGCAUCGUGCCGUCUCUUUCUCGG_CUUCUUGUUAACCCUCUCUUAC_CAAAC__CAUU ...((......))........(((.((((....(((((..(((((((((.....))))))))))))))...)))).))).......................... (-17.48 = -18.40 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:38 2006