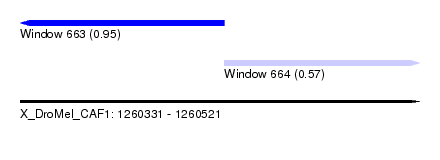
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,260,331 – 1,260,521 |
| Length | 190 |
| Max. P | 0.948847 |

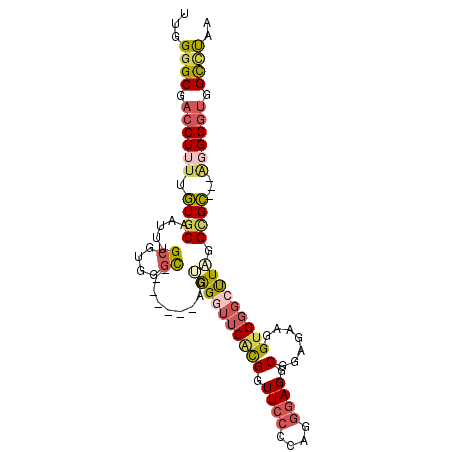
| Location | 1,260,331 – 1,260,428 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.23 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -18.50 |
| Energy contribution | -20.20 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1260331 97 - 22224390 AAGGGGAGGGUGGAGGUGGCGGCUU--CAGCGGGG------------------GGACUGCGAAUGAGGCAAGGCUGCGG-CGCUUGAUUGAUGGACUGCAGCGAAGGAGGAGCUGCCA ................(((((((((--(.((((..------------------...))))............(((((((-..((........)).)))))))......)))))))))) ( -35.10) >DroSec_CAF1 14523 96 - 1 AGAGGAGGG-GGGAGGUGGCGGCUU--CAGCGGGG------------------GAACUGCGAAUGAGGCAAGGCUGCGG-CGCUUGAGUGAUGGACUGCAGAGAAGGAGGAGCUGCCA .........-......(((((((((--(.((((..------------------...)))).............((((((-(((....))).....)))))).......)))))))))) ( -31.60) >DroSim_CAF1 13202 97 - 1 AGGGGAGGGGGGGAGGUGGCGGCUU--CAGCGGGG------------------GAACUGCGAAUGAGGCAAGGCUGCGG-CGCUUGAUUGAUGGACUGCAGCGAAGGAGGAGCUGCCA ................(((((((((--(.((((..------------------...))))............(((((((-..((........)).)))))))......)))))))))) ( -35.10) >DroEre_CAF1 16256 90 - 1 AAGUAGAGGG-------GGCGGCUU--CCGCGGGG------------------GGACUGCAAAUGAGGCAAGGCUGCGG-CGCUUGAUUGAUGGACUGCAGCGAAGGAGGAGCUGCCA .......((.-------(((..(((--((((((..------------------...))))............(((((((-..((........)).)))))))...))))).))).)). ( -34.60) >DroYak_CAF1 15803 108 - 1 ACGGGAAGAG-------GGCAGCUG--CCGCAAGGAGGGGGAGGUGAGGGGGUCGGCUGCAAAUGAGGCAAGGCUGCGG-CGCUUGAUUGAUGGACUGCAGCGUAGGAGGAGCUGUCG ..........-------(((((((.--((((........(.((.(((.....))).)).).......))...(((((((-..((........)).)))))))......))))))))). ( -33.16) >DroAna_CAF1 13764 82 - 1 AGGG------CUGGGGCGGCGGCCUUCCGGCAGGU------------------G-----C-----AGUUAAGAUUACGGACAGUUGAUUGAUUGACUGCGGCG--GGAAGAUGUGGCA ....------.....((.(((..(((((.((....------------------(-----(-----((((((((((((.....).)))))..)))))))).)).--))))).))).)). ( -28.80) >consensus AGGGGAAGGG_GGAGGUGGCGGCUU__CAGCGGGG__________________GGACUGCGAAUGAGGCAAGGCUGCGG_CGCUUGAUUGAUGGACUGCAGCGAAGGAGGAGCUGCCA .................((((((((....((((.......................))))............(((((((...((........)).))))))).......)))))))). (-18.50 = -20.20 + 1.70)

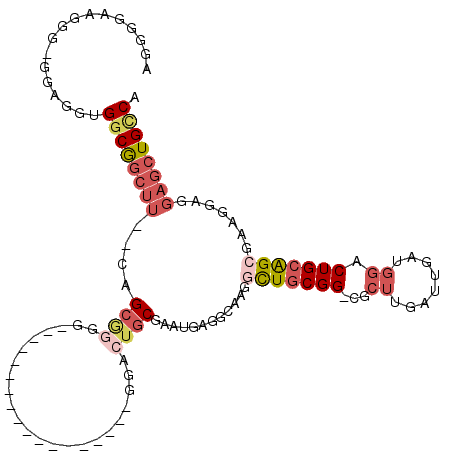
| Location | 1,260,428 – 1,260,521 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.66 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1260428 93 + 22224390 UUUGGGCGACCUUUUGUGCAAUUUGCGUUGGU-------AGUGGGUUCACGGUUCCCCAGGGAGGCGGAGAAGGUGGGCUUAGGUGC---AGGGUUGGUCUAA .(((((((((((..((..(...((((....))-------))(((((((((..((((((.....)).))))...))))))))).)..)---)))))).)))))) ( -30.90) >DroPse_CAF1 27579 73 + 1 UAGGGGCGACCUUUUAUGCAAUUUGCAUAGCC-------AGCAGCUGCUC-------UAGUG----------GGUG---CUGUGUGU---GUGGGCGGUUCAA ..(((.((.(((..(((((.....)))))(((-------((((.(..(..-------..)..----------).))---))).))..---..))))).))).. ( -25.80) >DroSec_CAF1 14619 93 + 1 UUGGAGCGACCUUUUGUGCAAUUUGCGUGGGC-------AGUGGGUUCACGGUUCCCCAGGGAGGCGGAGAAGGUGGGCUUAGGCGC---AGGGGUGGCCUAA .(((.((.((((((.((((...((((....))-------))(((((((((..((((((.....)).))))...))))))))).))))---)))))).))))). ( -37.30) >DroSim_CAF1 13299 93 + 1 UUGGGGCGACCUUUUGUGCAAUUUGCGUGGGC-------AGUGGGUUCACGGUUCCCCAGGGAGGCGGAGAAGGUGGGCUUAGGCGC---AGGGGUGGCCUAA ...((((.((((((.((((...((((....))-------))(((((((((..((((((.....)).))))...))))))))).))))---)))))).)))).. ( -40.20) >DroEre_CAF1 16346 98 + 1 UU-GGGCGACCUUUUGUGCAAUUUGCGUGGGCAGUGGGCAGUGGGUUCACGGUUCCCCAGGGAGGCAGAGAAGGUGGGCUUCUGCGCG----GGGUGGCCUAA .(-((((.((((..((((((.(((((.......((((((.....))))))..((((....)))))))))((((.....))))))))))----)))).))))). ( -40.00) >DroYak_CAF1 15911 96 + 1 UUGGGGCGACCUUUUGUGCAAUUUGCGUGGGC-------AGUGGCUUCGUGGUUCCCCAGGGAGGCAGAGACGGUGGGCUUUGGGGCGGCAGGGGUGGCCUAA ...((((.(((((...(((...(..((.((((-------....))))))..)..((((((((..((.......))...))))))))..)))))))).)))).. ( -34.80) >consensus UUGGGGCGACCUUUUGUGCAAUUUGCGUGGGC_______AGUGGGUUCACGGUUCCCCAGGGAGGCGGAGAAGGUGGGCUUAGGCGC___AGGGGUGGCCUAA ...((((.((((((.((((.....((....)).........((((((((((.((((....)))).).......))))))))).))))...)))))).)))).. (-19.56 = -20.66 + 1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:25 2006