
| Sequence ID | X_DroMel_CAF1 |
|---|---|
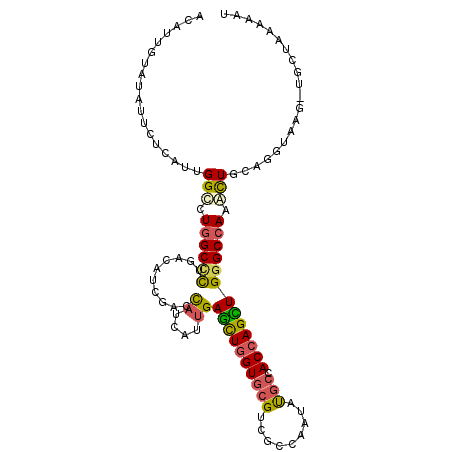
| Location | 11,320,830 – 11,320,935 |
| Length | 105 |
| Max. P | 0.618294 |

| Location | 11,320,830 – 11,320,935 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.35 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320830 105 + 22224390 ACAUUGUAUAUUCUUAUUGGCCUGGCCCUGACAUCGACCAUCAUUGAGCUGGUGCGAAGGCAAUAUGCCACCAGUUGGGCCAAGCUGCAGGUGGG-UUCUACAAGU ...(((((....(((((((((.((((((.(((.(((((((.(.....).)))).))).(((.....)))....))))))))).)))...))))))-...))))).. ( -35.10) >DroVir_CAF1 33063 105 + 1 ACGCUCUAUAUACUCAUUGGCCUGGCUCUAACCUCCACUAUCAUUGAGCUGGUGCGUCGCCAGUACGCGACCAGCUGGGCCAAGCUGCAGGUAAG-UGCUUACCUG ................(((((((((......)).............((((((((((((....).)))).))))))))))))))....(((((((.-...))))))) ( -36.70) >DroGri_CAF1 31527 105 + 1 ACUCUGUAUAUACUCAUUGGCCUGGCGCUAACGUCGACCAUUAUUGAGCUGGUACGUCGCCAGUACGCGACCAGCUGGGCCAAACUGCAGGUAUG-UGCUAACCCA .....(((((((((((((((((((((((....).)).)).......(((((((.((.((......))))))))))))))))))..))..))))))-)))....... ( -38.60) >DroEre_CAF1 20867 106 + 1 ACAUUGUAUAUUCUCAUUGGCUUGGCCUUGACAUCGACCAUUAUUGAGCUGGUGAGGCGGCAGUAUGCCACCAGCUGGGCCAAGCUGCAGGUAAGGUUCUACAAAU ...(((((...(((.(((((((((((((.....((((......))))((((((..((((......)))))))))).))))))))))...))).)))...))))).. ( -37.10) >DroWil_CAF1 35462 105 + 1 ACCUUAUACAUUUUGAUUGGUCUGGCCCUGACAUCGACCAUUAUAGAAUUGGUGCGUCGCCAAUAUGCCACAAGUUGGGCAAAAUUGCAGGUAAG-UGCAAAUAAU ((((....((.((((..((((.((((...(....)((((((((......))))).)))))))....)))))))).)).(((....)))))))...-.......... ( -21.00) >DroYak_CAF1 24191 102 + 1 ACAUUGUACAUCCUCAUUGGCCUGGCCCUGACAUCGACCAUCAUUGAGCUGGUGCGACGGCAAUAUGCCACCAGUUGGGCCAAACUGCAGGUAAGU----UCAAAU ...(((.((..(((((((((((..((.......(((((((.(.....).)))).))).(((.....)))....))..))))))..)).)))...))----.))).. ( -31.80) >consensus ACAUUGUAUAUUCUCAUUGGCCUGGCCCUGACAUCGACCAUCAUUGAGCUGGUGCGUCGCCAAUAUGCCACCAGCUGGGCCAAACUGCAGGUAAG_UGCUAAAAAU ..................(((.((((((..........((....))((((((((((.........))).))))))))))))).))).................... (-19.46 = -19.35 + -0.11)

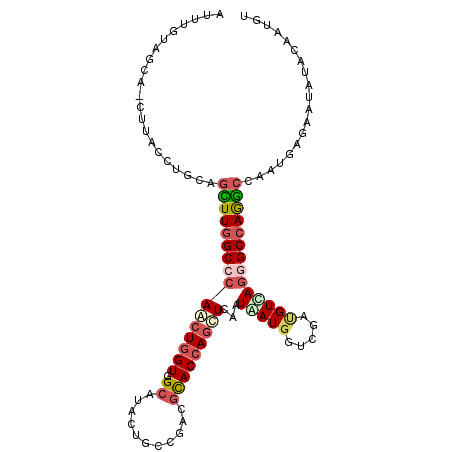
| Location | 11,320,830 – 11,320,935 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -19.53 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320830 105 - 22224390 ACUUGUAGAA-CCCACCUGCAGCUUGGCCCAACUGGUGGCAUAUUGCCUUCGCACCAGCUCAAUGAUGGUCGAUGUCAGGGCCAGGCCAAUAAGAAUAUACAAUGU ...(((((..-.....)))))(((((((((.......(((.....))).(((.((((.(.....).))))))).....)))))))))................... ( -34.30) >DroVir_CAF1 33063 105 - 1 CAGGUAAGCA-CUUACCUGCAGCUUGGCCCAGCUGGUCGCGUACUGGCGACGCACCAGCUCAAUGAUAGUGGAGGUUAGAGCCAGGCCAAUGAGUAUAUAGAGCGU (((((((...-.)))))))..((((((((.(((((((.((((.......))))))))))).............(((....))).)))))............))).. ( -36.50) >DroGri_CAF1 31527 105 - 1 UGGGUUAGCA-CAUACCUGCAGUUUGGCCCAGCUGGUCGCGUACUGGCGACGUACCAGCUCAAUAAUGGUCGACGUUAGCGCCAGGCCAAUGAGUAUAUACAGAGU .((((.....-...))))((...((((((.((((((((((((....))).)).)))))))......(((.((.(....))))))))))))...))........... ( -33.30) >DroEre_CAF1 20867 106 - 1 AUUUGUAGAACCUUACCUGCAGCUUGGCCCAGCUGGUGGCAUACUGCCGCCUCACCAGCUCAAUAAUGGUCGAUGUCAAGGCCAAGCCAAUGAGAAUAUACAAUGU ..(((((....((((..((..((((((((.....((((((.....))))))((((((.........)))).))......)))))))))).))))....)))))... ( -34.80) >DroWil_CAF1 35462 105 - 1 AUUAUUUGCA-CUUACCUGCAAUUUUGCCCAACUUGUGGCAUAUUGGCGACGCACCAAUUCUAUAAUGGUCGAUGUCAGGGCCAGACCAAUCAAAAUGUAUAAGGU ..........-...((((...((((((.......(.((((...((((((.((.((((.........)))))).)))))).)))).).....)))))).....)))) ( -23.80) >DroYak_CAF1 24191 102 - 1 AUUUGA----ACUUACCUGCAGUUUGGCCCAACUGGUGGCAUAUUGCCGUCGCACCAGCUCAAUGAUGGUCGAUGUCAGGGCCAGGCCAAUGAGGAUGUACAAUGU ..(((.----((...(((.(((((((((((.......(((.....)))((((.((((.(.....).))))))))....)))))))))...)))))..)).)))... ( -34.30) >consensus AUUUGUAGCA_CUUACCUGCAGCUUGGCCCAACUGGUGGCAUACUGCCGACGCACCAGCUCAAUAAUGGUCGAUGUCAGGGCCAGGCCAAUGAGAAUAUACAAUGU .....................((((((((((((((((.((...........)))))))))...(((((.....))))))))))))))................... (-19.53 = -20.57 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:33 2006