
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,320,341 – 11,320,491 |
| Length | 150 |
| Max. P | 0.993640 |

| Location | 11,320,341 – 11,320,451 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320341 110 + 22224390 AUAUACAAUAAUAACAUAGGCAUAAAAACACAAUCGUACACAGUGCGUAUGAGCGUUAAUAAUGCGACUGCAUUGAAAAGCAUUGCGCAUACGCACCGUUGGUUUUG---UGA ............................(((((......((.(((((((((.(((((..((((((....))))))...)))...)).))))))))).)).....)))---)). ( -27.40) >DroSec_CAF1 24453 109 + 1 AUAUACAAUAAUAACAUAGGAAUAAAAACAGAAUCGUACACAGUGCGUAUGAGCGAUAAUUAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGACUUU----UAA ..................((.............((((((.......))))))(((.....((((((..(((........)))...))))))))).)).........----... ( -20.10) >DroSim_CAF1 21244 109 + 1 AUAUACAAUAAUAACAUAGGAAUAAAAACAGAAGCGUACACAGUGCGUAUGAGCGAUAAUUAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGACUUU----UGA ............................((((((((...((.(((((((((.((.......((((....))))......))......))))))))).)))).))))----)). ( -23.72) >DroEre_CAF1 20402 105 + 1 UAACACAAUAAACACAUAG---UGAAAAUAAAAGC--GCACAGUGCGUAUGAGCGAUAAGAAUGCGGCUGGAUUAAAAAGCAUUACUCAUGCGCACCGUUGGUUUUU---UGA .....(((.((((.((..(---((..........)--))((.(((((((((((.......(((((..............))))).))))))))))).)))))))).)---)). ( -25.94) >DroYak_CAF1 23740 91 + 1 ----------------------UGAAAACAGAAGCGCGCACAGUGCGUAUGCGUGAUAUUAAUGCGGCUGCAUUCAAAAGCAUUACUCAUGCGCACCGUUGCUUUUUUUCUGA ----------------------......((((((.(((.((.(((((((((.(((((.(((((((....)))))...))..))))).))))))))).)))))....)))))). ( -29.80) >consensus AUAUACAAUAAUAACAUAGG_AUAAAAACAGAAGCGUACACAGUGCGUAUGAGCGAUAAUAAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGAUUUU____UGA .............................(((((((...((.(((((((((.(((.......))).(((.........)))......))))))))).)))))))))....... (-16.80 = -17.48 + 0.68)



| Location | 11,320,341 – 11,320,451 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320341 110 - 22224390 UCA---CAAAACCAACGGUGCGUAUGCGCAAUGCUUUUCAAUGCAGUCGCAUUAUUAACGCUCAUACGCACUGUGUACGAUUGUGUUUUUAUGCCUAUGUUAUUAUUGUAUAU .((---(((.....((((((((((((.((...(.....)(((((....)))))......)).))))))))))))......)))))....(((((..(((....))).))))). ( -30.10) >DroSec_CAF1 24453 109 - 1 UUA----AAAGUCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUAAUUAUCGCUCAUACGCACUGUGUACGAUUCUGUUUUUAUUCCUAUGUUAUUAUUGUAUAU ...----.......((((((((((((.(..(((.((....((((....)))))).)))..).))))))))))))(((((((...((...(((....)))..)).))))))).. ( -26.50) >DroSim_CAF1 21244 109 - 1 UCA----AAAGUCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUAAUUAUCGCUCAUACGCACUGUGUACGCUUCUGUUUUUAUUCCUAUGUUAUUAUUGUAUAU .((----.((((..((((((((((((.(..(((.((....((((....)))))).)))..).))))))))))))....)))).))............................ ( -26.50) >DroEre_CAF1 20402 105 - 1 UCA---AAAAACCAACGGUGCGCAUGAGUAAUGCUUUUUAAUCCAGCCGCAUUCUUAUCGCUCAUACGCACUGUGC--GCUUUUAUUUUCA---CUAUGUGUUUAUUGUGUUA ...---...(((((((((((((.(((((((((((..............)))))......)))))).))))))))((--((...........---....))))....)).))). ( -24.40) >DroYak_CAF1 23740 91 - 1 UCAGAAAAAAAGCAACGGUGCGCAUGAGUAAUGCUUUUGAAUGCAGCCGCAUUAAUAUCACGCAUACGCACUGUGCGCGCUUCUGUUUUCA---------------------- ........((((((..(((((((((((.((((((..(((....)))..))))))...))..((....))...)))))))))..))))))..---------------------- ( -27.70) >consensus UCA____AAAACCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUUAUUAUCGCUCAUACGCACUGUGUACGCUUCUGUUUUUAU_CCUAUGUUAUUAUUGUAUAU ..............((((((((((((.(((((((..............)))))......)).))))))))))))....................................... (-20.06 = -20.54 + 0.48)

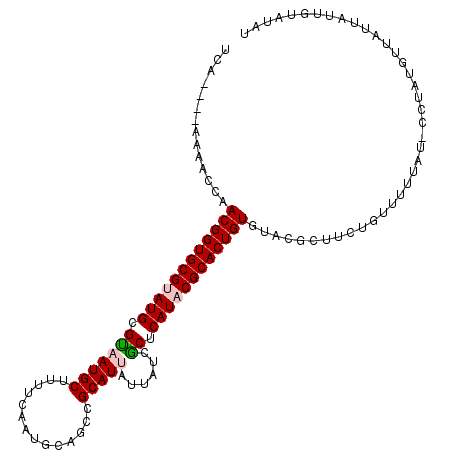
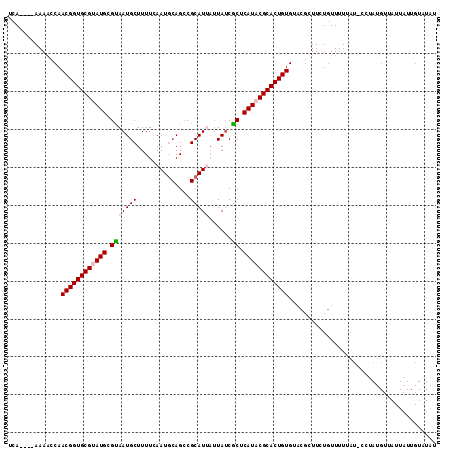
| Location | 11,320,374 – 11,320,491 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320374 117 + 22224390 UCGUACACAGUGCGUAUGAGCGUUAAUAAUGCGACUGCAUUGAAAAGCAUUGCGCAUACGCACCGUUGGUUUUG---UGAGCAUAUUUCAUUUGUGUAAUUAUUGCAAUGAUGCGUGAAG ...(((((((((((((((.(((((..((((((....))))))...)))...)).)))))))))..........(---((((.....))))).)))))).((((.(((....))))))).. ( -30.80) >DroSec_CAF1 24486 116 + 1 UCGUACACAGUGCGUAUGAGCGAUAAUUAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGACUUU----UAAGCAUAUUUCAUUUGUGCAAUUAUUGCAAUGAUGCGUGAAG .....(((.(((((((...((.......((((....))))......))..)))))))..(((.(((((.....----...(((((.......)))))........))))).))))))... ( -31.01) >DroSim_CAF1 21277 116 + 1 GCGUACACAGUGCGUAUGAGCGAUAAUUAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGACUUU----UGAGCAUAUUUCAUUUGUGCAAUUAUUGCAAUGAUGCGUGAAG (((((...(((.(((((((.......))))))))))((........))..))))).((((((.(((((.....----...(((((.......)))))........))))).))))))... ( -32.49) >DroEre_CAF1 20432 101 + 1 GC--GCACAGUGCGUAUGAGCGAUAAGAAUGCGGCUGGAUUAAAAAGCAUUACUCAUGCGCACCGUUGGUUUUU---UGAGCACAUUUCAUUUGAGU--------------UGUGUGUAG ((--((((((((((((((((.......(((((..............))))).))))))))))).(((.(.....---).)))...............--------------))))))).. ( -30.14) >DroYak_CAF1 23751 102 + 1 GCGCGCACAGUGCGUAUGCGUGAUAUUAAUGCGGCUGCAUUCAAAAGCAUUACUCAUGCGCACCGUUGCUUUUUUUCUGAGCAUACUUCAUUUG-----------------UGUGUUUA- ..((((((((((((((((.(((((.(((((((....)))))...))..))))).))))))))).(((((((.......))))).))......))-----------------)))))...- ( -34.20) >consensus GCGUACACAGUGCGUAUGAGCGAUAAUAAUGCGGCUGCAUUGAAAAGCAUUACGCAUACGCACCGUUGAUUUU____UGAGCAUAUUUCAUUUGUGCAAUUAUUGCAAUGAUGCGUGAAG .....(((.(((((((((.(((.......))).(((.........)))......))))))))).................(((((.......))))).................)))... (-18.72 = -19.20 + 0.48)

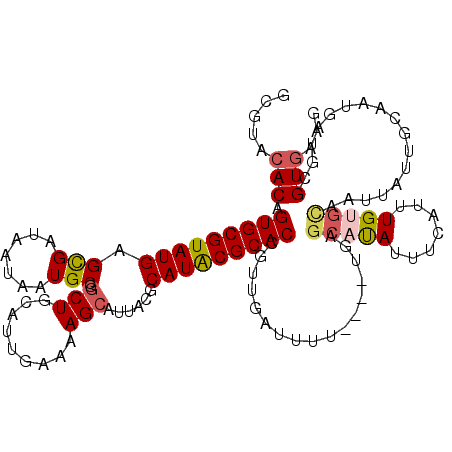

| Location | 11,320,374 – 11,320,491 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11320374 117 - 22224390 CUUCACGCAUCAUUGCAAUAAUUACACAAAUGAAAUAUGCUCA---CAAAACCAACGGUGCGUAUGCGCAAUGCUUUUCAAUGCAGUCGCAUUAUUAACGCUCAUACGCACUGUGUACGA ......(((....)))......(((((...(((.......)))---..........((((((((((.((...(.....)(((((....)))))......)).)))))))))))))))... ( -28.20) >DroSec_CAF1 24486 116 - 1 CUUCACGCAUCAUUGCAAUAAUUGCACAAAUGAAAUAUGCUUA----AAAGUCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUAAUUAUCGCUCAUACGCACUGUGUACGA ......(((((((((((.....))))...))))....)))...----...((..((((((((((((.(..(((.((....((((....)))))).)))..).))))))))))))..)).. ( -31.00) >DroSim_CAF1 21277 116 - 1 CUUCACGCAUCAUUGCAAUAAUUGCACAAAUGAAAUAUGCUCA----AAAGUCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUAAUUAUCGCUCAUACGCACUGUGUACGC ......(((((((((((.....))))...))))....)))...----...((..((((((((((((.(..(((.((....((((....)))))).)))..).))))))))))))..)).. ( -31.00) >DroEre_CAF1 20432 101 - 1 CUACACACA--------------ACUCAAAUGAAAUGUGCUCA---AAAAACCAACGGUGCGCAUGAGUAAUGCUUUUUAAUCCAGCCGCAUUCUUAUCGCUCAUACGCACUGUGC--GC .....((((--------------..((....))..))))....---........((((((((.(((((((((((..............)))))......)))))).))))))))..--.. ( -25.34) >DroYak_CAF1 23751 102 - 1 -UAAACACA-----------------CAAAUGAAGUAUGCUCAGAAAAAAAGCAACGGUGCGCAUGAGUAAUGCUUUUGAAUGCAGCCGCAUUAAUAUCACGCAUACGCACUGUGCGCGC -........-----------------.........................((.(((((((((.(((.((((((..(((....)))..))))))...))).))....)))))))..)).. ( -24.00) >consensus CUUCACGCAUCAUUGCAAUAAUUACACAAAUGAAAUAUGCUCA____AAAACCAACGGUGCGUAUGCGUAAUGCUUUUCAAUGCAGCCGCAUUAUUAUCGCUCAUACGCACUGUGUACGC ......................................................((((((((((((.(((((((..............)))))......)).))))))))))))...... (-20.06 = -20.54 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:30 2006