| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,314,630 – 11,314,721 |
| Length | 91 |
| Max. P | 0.853082 |

| Location | 11,314,630 – 11,314,721 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 57.19 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -9.08 |
| Energy contribution | -12.61 |
| Covariance contribution | 3.53 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11314630 91 - 22224390 GGGCAUGGGCUGGGCAUGGUGGUGGUU------------CCAGAGAUUGUCAUCGUUAUCAUCGGGUCUA-------GACCCAUUCAGCCACAUGGACAGGAUGAGCACU (..(((((((((((.(((((((..(((------------.....)))..))))))).......((((...-------.)))).))))))).))))..)............ ( -31.40) >DroPse_CAF1 25251 88 - 1 G------GACUGGGCACAGACCCAGU-------------UCAGAGAUUGUGAUGAUUA---UCAGGUCUAGACAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN (------(((((((......))))))-------------))..(((((.((((....)---))))))))......................................... ( -17.20) >DroSec_CAF1 17724 93 - 1 G------GGCUGGGCAUGGUGGUAGCU-----------ACCAGAGAUUGCCAUCGUUAUCAUCGGGUCUAGACUCUAGACCCAUUCAGCCACAUGGCCAGGAUGAGCACU .------......((((((((((((((-----------.....)).))))))))))..(((((((((((((...)))))))).....(((....)))...)))))))... ( -35.90) >DroSim_CAF1 14863 93 - 1 G------GGCUGGGCAUGGUGGUAGCU-----------ACCAGAGAUUGCCAUCGCUAUCAUCGGGUCUAGACUCUAGACCCAUUCAGCCACAUGGCCAGGAUGAGCACU .------.((..(((((((..((..(.-----------....)..))..)))).))).(((((((((((((...)))))))).....(((....)))...)))))))... ( -37.10) >DroYak_CAF1 18614 93 - 1 G------GACUGGGCGUGGUGGUGAUUCGCUGGCAAACGCCAGUGAUUGCCAUCAUUG---UCGGGUCUG-------GACACAGCCAGUCACAUGGCCA-GAUGACCACG (------((((.((((((((((..((.(((((((....)))))))))..)))))).))---)).)))))(-------(.(((.((((......))))..-).)).))... ( -44.80) >DroPer_CAF1 32594 88 - 1 G------GACUGGGCACAGACCCAGU-------------UCAGAGAUUGUGAUGAUUA---UCAGGUCUAGACAAUGGACCCCCCACACACACACACUCGGACACACACU (------(((((((......))))))-------------)).(((..((((.((....---...((((((.....)))))).......))))))..)))........... ( -27.74) >consensus G______GACUGGGCAUGGUGGUAGUU____________CCAGAGAUUGCCAUCAUUA___UCGGGUCUAGAC____GACCCAUUCAGCCACAUGGCCAGGAUGAGCACU .......((((((..((((((((((((.................)))))))))))).......(((((.........)))))..)))))).................... ( -9.08 = -12.61 + 3.53)

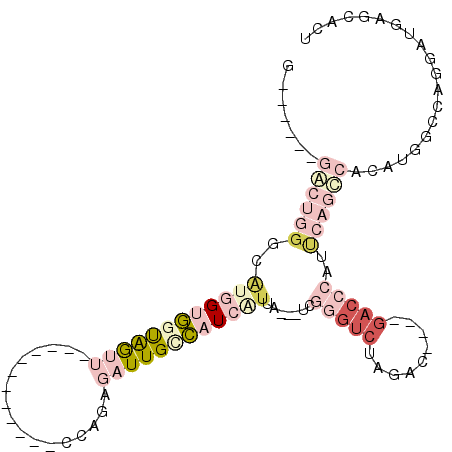

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:25 2006