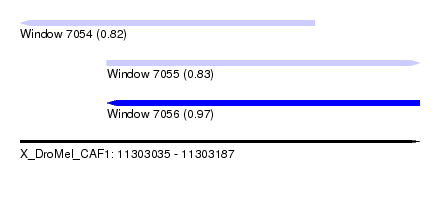
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,303,035 – 11,303,187 |
| Length | 152 |
| Max. P | 0.969631 |

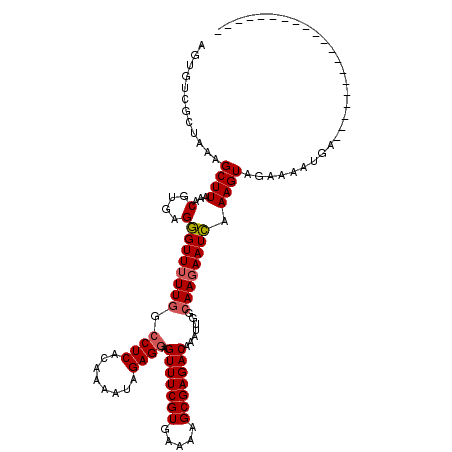
| Location | 11,303,035 – 11,303,147 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11303035 112 - 22224390 AAAAUAGAGAAGUUUCGUGAAAAGCGAGACUACUUGGCAAGAAUUAAAGUAGAAAAUGACGUACUUUACUGUCUUGAAGACUUAACAGUCCCCACUUGUACUCCCUAAGUAU ..........((((((((.....))))))))(((((((((((..(((((((..........)))))))...)))))..((((....))))..............)))))).. ( -24.80) >DroSec_CAF1 6578 91 - 1 AAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGAAAAUAAUCAAAGUAGAAAAUAA---------------------CCUAACAGUCCCUACUUCAACUGACUAAUUAU ....((((((.(((((((.....)))))))...((((......))))............---------------------)))..((((..........)))).)))..... ( -15.10) >DroSim_CAF1 7633 91 - 1 AAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGGCAAGAAUCAAAGUAGAAAAUGA---------------------CCUAACAGUCCCUACUUCAUCUCACUAAUUAU ...........(((((((.....))))))).((((((..(((....((((((.....((---------------------(......))).))))))..)))..)))))).. ( -18.50) >consensus AAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGGCAAGAAUCAAAGUAGAAAAUGA_____________________CCUAACAGUCCCUACUUCAACUCACUAAUUAU ...........(((((((.....))))))).((((((...((....((((((.......................................))))))....)).)))))).. (-11.45 = -11.23 + -0.22)



| Location | 11,303,068 – 11,303,187 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11303068 119 + 22224390 CUUCAAGACAGUAAAGUACGUCAUUUUCUACUUUAAUUCUUGCCAAGUAGUCUCGCUUUUCACGAAACUUCUCUAUUUUGUGAGGCCAAAAACUCUUACGUUAAAGCUCUAGUGACACU ...(((((...(((((((.(......).)))))))..)))))......(((.(((((..((((((((........))))))))(((....(((......)))...)))..))))).))) ( -21.30) >DroSec_CAF1 6610 99 + 1 --------------------UUAUUUUCUACUUUGAUUAUUUUCAAUUUGUCUCGCUUUUCACGAAACUCCUCUAUUUUGUGAGGCCAAAAACCCUCACGUUUAAGCUUUAGCGACACU --------------------............((((......))))...((.(((((.......((((...........((((((........)))))))))).......))))).)). ( -17.14) >DroSim_CAF1 7665 99 + 1 --------------------UCAUUUUCUACUUUGAUUCUUGCCAAUUUGUCUCGCUUUUCACGAAACUCCUCUAUUUUGUGAGGCCAAAAACCCUCACGUUUAAGCUUUAGCGACACU --------------------.............................((.(((((.......((((...........((((((........)))))))))).......))))).)). ( -15.74) >consensus ____________________UCAUUUUCUACUUUGAUUCUUGCCAAUUUGUCUCGCUUUUCACGAAACUCCUCUAUUUUGUGAGGCCAAAAACCCUCACGUUUAAGCUUUAGCGACACU .................................................((.(((((..((((((((........))))))))(((...((((......))))..)))..))))).)). (-14.42 = -14.53 + 0.11)

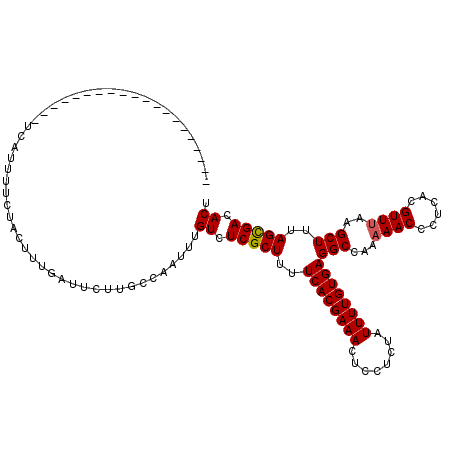
| Location | 11,303,068 – 11,303,187 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11303068 119 - 22224390 AGUGUCACUAGAGCUUUAACGUAAGAGUUUUUGGCCUCACAAAAUAGAGAAGUUUCGUGAAAAGCGAGACUACUUGGCAAGAAUUAAAGUAGAAAAUGACGUACUUUACUGUCUUGAAG .(((...((((((((((......)))).))))))...)))......(((.((((((((.....)))))))).)))..(((((..(((((((..........)))))))...)))))... ( -28.40) >DroSec_CAF1 6610 99 - 1 AGUGUCGCUAAAGCUUAAACGUGAGGGUUUUUGGCCUCACAAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGAAAAUAAUCAAAGUAGAAAAUAA-------------------- .((...((....))....)).(((..(((((..(((((........)))).(((((((.....)))))))...)..)))))..))).............-------------------- ( -22.00) >DroSim_CAF1 7665 99 - 1 AGUGUCGCUAAAGCUUAAACGUGAGGGUUUUUGGCCUCACAAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGGCAAGAAUCAAAGUAGAAAAUGA-------------------- ......(((((.........((((((.(....).))))))...........(((((((.....)))))))...))))).....................-------------------- ( -24.20) >consensus AGUGUCGCUAAAGCUUAAACGUGAGGGUUUUUGGCCUCACAAAAUAGAGGAGUUUCGUGAAAAGCGAGACAAAUUGGCAAGAAUCAAAGUAGAAAAUGA____________________ ............((((...(....)((((((((.((((........)))).(((((((.....))))))).......)))))))).))))............................. (-18.41 = -18.97 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:23 2006