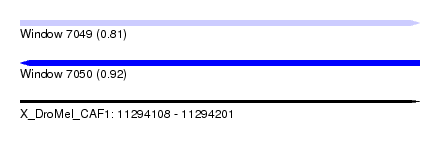
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,294,108 – 11,294,201 |
| Length | 93 |
| Max. P | 0.921158 |

| Location | 11,294,108 – 11,294,201 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.05 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -7.74 |
| Energy contribution | -8.72 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11294108 93 + 22224390 CAUUUGUUGUCUUGCUAAUUU---GCGCAC---GCAC--ACACGCCGCAUUUGCA--UUUGCAUUUGCAUCUGCAAUAUGCAUU-------UGCAUCUUUUAAUGCAAC--G .....(((((.(((......(---(((...---((..--....))))))..((((--..(((((((((....)))).)))))..-------)))).....))).)))))--. ( -25.40) >DroPse_CAF1 26180 104 + 1 CAGUUGUUUUAUUGCUAAUUUUAAGCGCUCAGUGCAUUCACGAGGCGCAUUUUGAGGUUUGA------GUCAGUGUUUGGUA--CAGUCCCGGCGGCAUUUGGUAAAUCUCA ........................(((((..(((....)))..)))))....(((((((((.------...(((((((((..--.....)))..))))))...))))))))) ( -23.90) >DroSec_CAF1 24280 93 + 1 CAUUUGUUGUCUUGCUAAUUU---GCGCAC---GCAC--ACACGCCGCAUUUGCA--UUUGCAUUUGCAUCUGCAAUUUGCAUU-------UGCAUCUUUUAAUGCAAC--G .....(((((.(((......(---(((...---((..--....))))))..((((--..((((.((((....))))..))))..-------)))).....))).)))))--. ( -24.50) >DroSim_CAF1 19409 93 + 1 CAUUUGUUGUCUUGCUAAUUU---GCGCAC---GCAC--ACACGCCGCAUUUGCA--UUUGCAUUUGCAUCUGCAAUUUGCAUU-------UGCAUCUUUUAAUGCAAC--G .....(((((.(((......(---(((...---((..--....))))))..((((--..((((.((((....))))..))))..-------)))).....))).)))))--. ( -24.50) >DroEre_CAF1 24011 81 + 1 CAUUUGCUGUCUUGCUAAUUU---GCGCAC---GCAC--ACACGCCGCAUU--------------UGCAUCUGCAAUUUGCAUU-------UGCAUCUUUUAAUGCAUC--C ....((((((...((......---))))).---))).--.......(((((--------------((((..(((.....)))..-------))))......)))))...--. ( -16.90) >DroYak_CAF1 24775 83 + 1 CAUUUGCUGUCUUGCUAAUUU---GCGCAC---GCAC--ACACGCCGCAUU--------------UGCAUCUGCAAUUUGCAUU-------UGCAUCUUUUAAUGCAUCUUU ....((((((...((......---))))).---))).--.......(((((--------------((((..(((.....)))..-------))))......)))))...... ( -16.90) >consensus CAUUUGUUGUCUUGCUAAUUU___GCGCAC___GCAC__ACACGCCGCAUUUGCA__UUUGC___UGCAUCUGCAAUUUGCAUU_______UGCAUCUUUUAAUGCAAC__G ....(((....((((...........((.....))...........(((..(((......)))..)))....))))...))).........(((((......)))))..... ( -7.74 = -8.72 + 0.97)

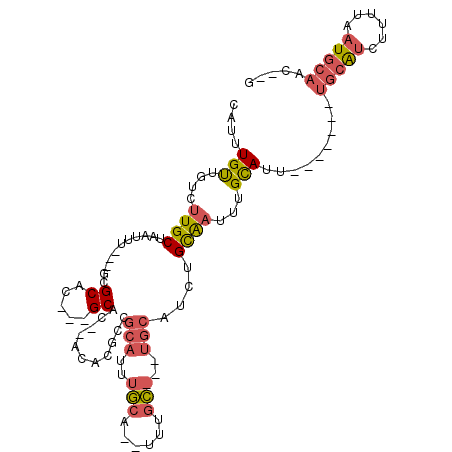
| Location | 11,294,108 – 11,294,201 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.05 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -8.72 |
| Energy contribution | -9.97 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11294108 93 - 22224390 C--GUUGCAUUAAAAGAUGCA-------AAUGCAUAUUGCAGAUGCAAAUGCAAA--UGCAAAUGCGGCGUGU--GUGC---GUGCGC---AAAUUAGCAAGACAACAAAUG (--((((((((......((((-------..(((((.((((....)))))))))..--)))))))))))))(((--...(---.(((..---......))).)...))).... ( -28.00) >DroPse_CAF1 26180 104 - 1 UGAGAUUUACCAAAUGCCGCCGGGACUG--UACCAAACACUGAC------UCAAACCUCAAAAUGCGCCUCGUGAAUGCACUGAGCGCUUAAAAUUAGCAAUAAAACAACUG ..............(((....((.....--..))..........------..............((((.(((((....))).)))))).........)))............ ( -14.00) >DroSec_CAF1 24280 93 - 1 C--GUUGCAUUAAAAGAUGCA-------AAUGCAAAUUGCAGAUGCAAAUGCAAA--UGCAAAUGCGGCGUGU--GUGC---GUGCGC---AAAUUAGCAAGACAACAAAUG .--.(((((((....))))))-------).(((...(((((..(((....)))..--))))).((((((((..--..))---)).)))---).....)))............ ( -27.70) >DroSim_CAF1 19409 93 - 1 C--GUUGCAUUAAAAGAUGCA-------AAUGCAAAUUGCAGAUGCAAAUGCAAA--UGCAAAUGCGGCGUGU--GUGC---GUGCGC---AAAUUAGCAAGACAACAAAUG .--.(((((((....))))))-------).(((...(((((..(((....)))..--))))).((((((((..--..))---)).)))---).....)))............ ( -27.70) >DroEre_CAF1 24011 81 - 1 G--GAUGCAUUAAAAGAUGCA-------AAUGCAAAUUGCAGAUGCA--------------AAUGCGGCGUGU--GUGC---GUGCGC---AAAUUAGCAAGACAGCAAAUG .--..((((((....))))))-------..(((...((((...(((.--------------...)))(((..(--....---)..)))---......))))....))).... ( -21.20) >DroYak_CAF1 24775 83 - 1 AAAGAUGCAUUAAAAGAUGCA-------AAUGCAAAUUGCAGAUGCA--------------AAUGCGGCGUGU--GUGC---GUGCGC---AAAUUAGCAAGACAGCAAAUG .....((((((....))))))-------..(((...((((...(((.--------------...)))(((..(--....---)..)))---......))))....))).... ( -21.20) >consensus C__GAUGCAUUAAAAGAUGCA_______AAUGCAAAUUGCAGAUGCA___GCAAA__UGCAAAUGCGGCGUGU__GUGC___GUGCGC___AAAUUAGCAAGACAACAAAUG .....((((((..((..((((.........))))..))...))))))................(((.........((((.....)))).........)))............ ( -8.72 = -9.97 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:18 2006