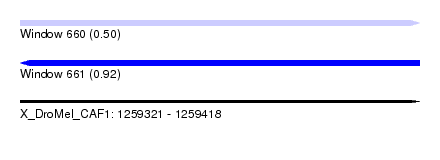
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,259,321 – 1,259,418 |
| Length | 97 |
| Max. P | 0.916861 |

| Location | 1,259,321 – 1,259,418 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -16.81 |
| Energy contribution | -15.63 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1259321 97 + 22224390 ----------UUCCAACUUGUACAGUUGGCUUAU---UAGCCAUAUUAUU-----AUCUCCUAUUAGCCCGAUUUCGGCGU-UUGCCUUUAAGCGAGCUGCAUUGCAGCGCACUGA ----------............(((((((((...---.))))).......-----...........(((((....)))(((-(((....)))))).(((((...))))))))))). ( -24.50) >DroSec_CAF1 13520 94 + 1 ----------UUCCAACUGGUG---CUGGCUUAU---AAGCCAUAUUAUU-----AUCUUCUAUUAGCCCGAUUUCGGCGU-UUGCCUUUAAGCGAGCUGCAUUGCAGCGCACUGA ----------........((((---((((((...---.))))).......-----...........(((((....))).))-((((......))))(((((...)))))))))).. ( -25.60) >DroSim_CAF1 12182 94 + 1 ----------UUCCAACUGGUG---CUGGCUUAU---AAGCCAUAUUAUU-----AUCUUCUAUUAGCCCGAUUUCGGCGU-UUGCCUUUAAGCGAGCUGCAUUGCAGCGCACUGG ----------......(..(((---((((((...---.))))).......-----...........(((((....))).))-((((......))))(((((...)))))))))..) ( -27.80) >DroEre_CAF1 15316 110 + 1 UUGUGAAUUGUUCCAACUGCUC---U-GGCUAAUUUAGAGCCGCAUUAAUCUCCAAUCUCCUAUUAGCCUGAU--GUCGGCGCUGCCUUUAGGCCAGCUGCAGUGCAGCGCAGCGA (((((...(((....(((((.(---(-((((((..(((.(((((((((..((.............))..))))--).)))).)))...))).)))))..)))))))).)))))... ( -31.92) >consensus __________UUCCAACUGGUG___CUGGCUUAU___AAGCCAUAUUAUU_____AUCUCCUAUUAGCCCGAUUUCGGCGU_UUGCCUUUAAGCGAGCUGCAUUGCAGCGCACUGA .................((((......((((.......))))........................((..........(((.(((....)))))).(((((...))))))))))). (-16.81 = -15.63 + -1.19)

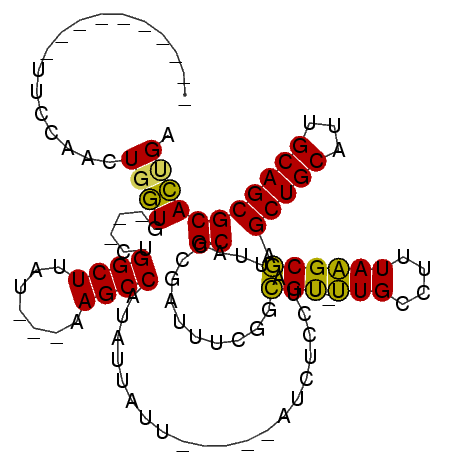
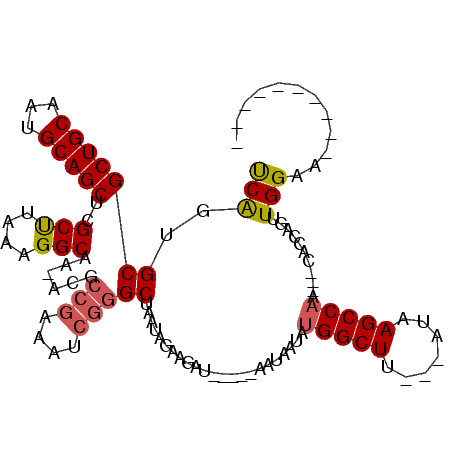
| Location | 1,259,321 – 1,259,418 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1259321 97 - 22224390 UCAGUGCGCUGCAAUGCAGCUCGCUUAAAGGCAA-ACGCCGAAAUCGGGCUAAUAGGAGAU-----AAUAAUAUGGCUA---AUAAGCCAACUGUACAAGUUGGAA---------- ((((((.(((((...))))).))))....(((..-..))))).....(((((.((..(...-----..)..))))))).---.....((((((.....))))))..---------- ( -26.30) >DroSec_CAF1 13520 94 - 1 UCAGUGCGCUGCAAUGCAGCUCGCUUAAAGGCAA-ACGCCGAAAUCGGGCUAAUAGAAGAU-----AAUAAUAUGGCUU---AUAAGCCAG---CACCAGUUGGAA---------- ...((((((......))((((((......(((..-..))).....))))))..........-----.......(((((.---...))))))---))).........---------- ( -26.40) >DroSim_CAF1 12182 94 - 1 CCAGUGCGCUGCAAUGCAGCUCGCUUAAAGGCAA-ACGCCGAAAUCGGGCUAAUAGAAGAU-----AAUAAUAUGGCUU---AUAAGCCAG---CACCAGUUGGAA---------- (((((((((......))((((((......(((..-..))).....))))))..........-----.......(((((.---...))))))---)))....)))..---------- ( -27.80) >DroEre_CAF1 15316 110 - 1 UCGCUGCGCUGCACUGCAGCUGGCCUAAAGGCAGCGCCGAC--AUCAGGCUAAUAGGAGAUUGGAGAUUAAUGCGGCUCUAAAUUAGCC-A---GAGCAGUUGGAACAAUUCACAA .((((..(((((...)))))..(((....)))))))(((((--.((.(((((((.((((.(((.(......).)))))))..)))))))-.---))...)))))............ ( -36.20) >consensus UCAGUGCGCUGCAAUGCAGCUCGCUUAAAGGCAA_ACGCCGAAAUCGGGCUAAUAGAAGAU_____AAUAAUAUGGCUU___AUAAGCCAA___CACCAGUUGGAA__________ (((..(((((((...)))))..(((....)))......(((....))))).......................(((((.......)))))...........)))............ (-17.19 = -17.62 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:23 2006