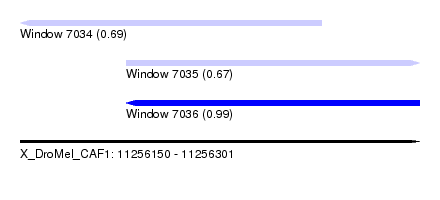
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,256,150 – 11,256,301 |
| Length | 151 |
| Max. P | 0.985751 |

| Location | 11,256,150 – 11,256,264 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -40.63 |
| Consensus MFE | -31.86 |
| Energy contribution | -31.42 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11256150 114 - 22224390 UGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC------CACCUUCGGGGGAUUCUCGUGACCGAAAUGGACCAGGGAAA .(((.((.((...(((((((((((((.((......))..))))))))))))))))).))).....(((((..(------((..(((((.............))))).)))...))))).. ( -41.92) >DroSec_CAF1 15528 111 - 1 UGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCA---------CCUUGGAGGGCUCCUCGUAACCGAAAUGGACCAGGGAAA .(((.((.((...(((((((((((((.((......))..))))))))))))))))).)))............---------(((((((((...))))....((.....))..)))))... ( -39.90) >DroSim_CAF1 16164 111 - 1 UAGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAUCUUGGUCUGCAUCCUGGAGAUCCGUACAUCCUUCA---------CCUUGCAGGGCUCCUCGUGACCGAAAUGGACCAGGGAAA ..(((((.(.((((((((((((((((.............))))))))))))(((((...........)))))---------..)))).))))))(((....((.....))....)))... ( -36.72) >DroEre_CAF1 15300 111 - 1 UGGAGCUGCAGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCACAU---------UGGCCUGCUCCUCGUGGCUGAAAUGGACCAGGGUGA .(((.((.((....((((((((((((.((......))..)))))))))))))).)).)))...((((((((.(((---------(((((.((.....))))))..))))))..)))))). ( -42.90) >DroYak_CAF1 15945 120 - 1 CGGAGCCGCAGCAAGGAUGCAGGCCAUCGAUGUCCAGUUUGGUCUGCAUCCUGGAAAUCCUCACAUCCCUCACAUCCCUCACCUUCGAGUGCUCCUCGUGGCCGAAAUGGACCAGGGAAA .(((.((......(((((((((((((.(........)..)))))))))))))))...))).....(((((....(((.((.((..((((.....)))).))..))...)))..))))).. ( -41.70) >consensus UGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC________CCUUGGAGGGCUCCUCGUGACCGAAAUGGACCAGGGAAA ..(((..(((...(((((((((((((.............)))))))))))))(((..........))).....................)))..)))....((...........)).... (-31.86 = -31.42 + -0.44)


| Location | 11,256,190 – 11,256,301 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11256190 111 + 22224390 G------GUGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUUGCCAACACUGACAGC---CCGCAGUCAUC (------(..(((((..((((((.(((((((((((((.(((..(((....))).))).)))))))))...)).)).))).)))..)))))..)).....((((...---.....)))).. ( -42.10) >DroSec_CAF1 15567 109 + 1 --------UGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAGCUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUCGCCAGCACUGACAAC---CCGCCGUCCUC --------.((((((..((((((.(((((((((((((.(((..(((....))).))).)))))))))...)).)).))).)))..)))))).........(((...---.....)))... ( -36.60) >DroSim_CAF1 16203 109 + 1 --------UGAAGGAUGUACGGAUCUCCAGGAUGCAGACCAAGAUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCUAUUGAUUCCUUCGCCAGCACUGACAAC---CCGCAGUCCAA --------....((((....((....))(((((((((.(((..((....))...))).)))))))))((((((((..((..(((.....)))..))..))).....---))))))))).. ( -33.70) >DroEre_CAF1 15336 112 + 1 -----AUGUGAAGGAUGUGAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCUGCAGCUCCAUUGAUUCCCUUGCCAUCAGUGGCAAG---CCGCAGUCCUC -----.............(((((.....(((((((((.(((..(((....))).))).)))))))))..((((.(((((((((((.........))))))))..))---).))))))))) ( -41.50) >DroYak_CAF1 15985 117 + 1 GAGGGAUGUGAGGGAUGUGAGGAUUUCCAGGAUGCAGACCAAACUGGACAUCGAUGGCCUGCAUCCUUGCUGCGGCUCCGUUGAUUCCCUGGCC---AGUGGCAAGAAACCAAAGUCCUA ..(((((((.(((((.....(((.....(((((((((.(((.............))).))))))))).((....)))))......))))).)).---..(((.......)))..))))). ( -38.62) >consensus _______GUGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUCGCCAGCACUGACAAC___CCGCAGUCCUC .......((((((((..((((((.(((((((((((((.(((.............))).)))))))))...)).)).))).)))..)))))))).......(((...........)))... (-29.16 = -29.20 + 0.04)



| Location | 11,256,190 – 11,256,301 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -34.08 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11256190 111 - 22224390 GAUGACUGCGG---GCUGUCAGUGUUGGCAAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC------C ((((..(((.(---((.......))).)))(((((.((......((....)).(((((((((((((.((......))..)))))))))))))...))))))).))))......------. ( -40.20) >DroSec_CAF1 15567 109 - 1 GAGGACGGCGG---GUUGUCAGUGCUGGCGAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCA-------- .......((.(---((.......))).))((((((......(((.((.((...(((((((((((((.((......))..))))))))))))))))).))).....)))))).-------- ( -43.40) >DroSim_CAF1 16203 109 - 1 UUGGACUGCGG---GUUGUCAGUGCUGGCGAAGGAAUCAAUAGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAUCUUGGUCUGCAUCCUGGAGAUCCGUACAUCCUUCA-------- ..(((.(((((---(((..(..(((.((((......((....))..)))))))(((((((((((((.............))))))))))))))..))))))))..)))....-------- ( -42.82) >DroEre_CAF1 15336 112 - 1 GAGGACUGCGG---CUUGCCACUGAUGGCAAGGGAAUCAAUGGAGCUGCAGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCACAU----- (((((((((((---((..(((.((((.........)))).)))))))))))..(((((((((((((.((......))..))))))))))))).....))))).............----- ( -49.60) >DroYak_CAF1 15985 117 - 1 UAGGACUUUGGUUUCUUGCCACU---GGCCAGGGAAUCAACGGAGCCGCAGCAAGGAUGCAGGCCAUCGAUGUCCAGUUUGGUCUGCAUCCUGGAAAUCCUCACAUCCCUCACAUCCCUC ..(((..(((((((((((((...---)).))))))))))).(((((....)).(((((((((((((.(........)..)))))))))))))((....)).....)))......)))... ( -41.40) >consensus GAGGACUGCGG___GUUGUCAGUGCUGGCAAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC_______ (((((((((((....((((((....)))))).....((....)).))))))..(((((((((((((.............))))))))))))).....))))).................. (-34.08 = -34.60 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:05 2006