
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,248,965 – 11,249,111 |
| Length | 146 |
| Max. P | 0.993814 |

| Location | 11,248,965 – 11,249,085 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
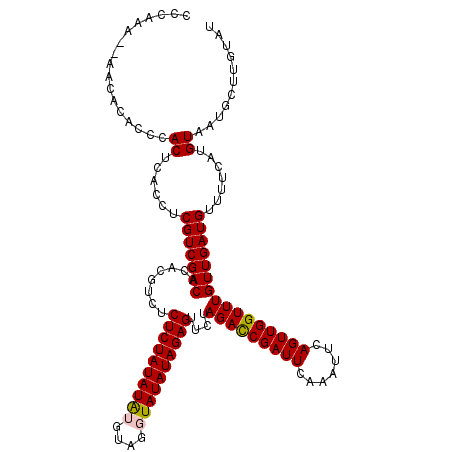
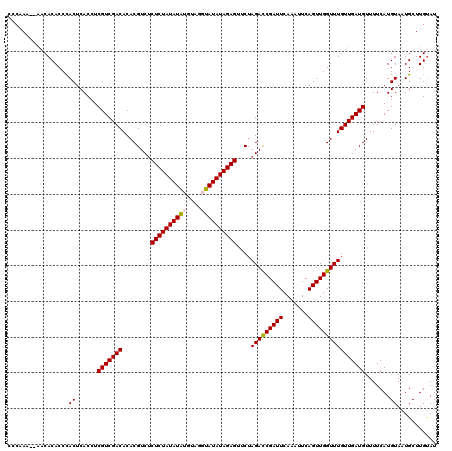
>X_DroMel_CAF1 11248965 120 + 22224390 CCCAAAAAAACACACCCACUCACCUCGUCGACACACGUCUCUCUAUAUAUGUAGGUAUAUAGAGUACUAGAUCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGCAU .........................(((((((....((..((((((((((....)))))))))).)).(((((((((.......)))))))))))))))).....(((((.....))))) ( -23.60) >DroSec_CAF1 8398 118 + 1 CCCAAA--AACACACCCACUCACCUCGUCGACACACGUCUCUCUAUAUAUGUAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAU ......--...(((..((..((...(((((((........((((((((((....))))))))))....(((((((((.......))))))))))))))))......))...))..))).. ( -24.60) >DroSim_CAF1 9055 118 + 1 CCCAAA--AACACACCCACUCACCUCGUCGACACACGUCUCUCUAUAUAUGCAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAU ......--...(((..((..((...(((((((........((((((((((....))))))))))....(((((((((.......))))))))))))))))......))...))..))).. ( -24.60) >DroEre_CAF1 8333 116 + 1 CCCAAU----CACACCUACUCACCUCGUCGACACACGUCUCUCUAUAUGAAUACAUAUAUAGAGUUCUAGACCGAUUCACAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAUUGCUUGUAU ......----......(((......(((((((......((((.((((((....)))))).))))....(((((((((.......)))))))))))))))).......))).......... ( -23.72) >DroYak_CAF1 8436 117 + 1 CCCAAA-CAACACACCUACUCACCUCGUCGACACACGUCCCUCUAUAUAAAUA--UAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUACUGCUUGUAU .....(-(((((....(((......(((((((........(((((((((....--)))))))))....(((((((((.......)))))))))))))))).......))).)).)))).. ( -25.32) >consensus CCCAAA__AACACACCCACUCACCUCGUCGACACACGUCUCUCUAUAUAUGUAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAU .................((......(((((((........((((((((((....))))))))))....(((((((((.......)))))))))))))))).......))........... (-22.88 = -22.96 + 0.08)

| Location | 11,249,005 – 11,249,111 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
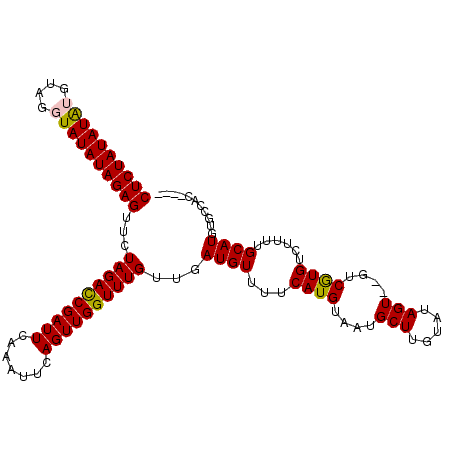
>X_DroMel_CAF1 11249005 106 + 22224390 CUCUAUAUAUGUAGGUAUAUAGAGUACUAGAUCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGCAUAGU--GUCGUGUCUUUUGCAUGUGCCAC---- ((((((((((....))))))))))...((((((((((.......)))))))))).....((...(((((((.....((((...--...))))...))))))).))...---- ( -22.20) >DroSec_CAF1 8436 108 + 1 CUCUAUAUAUGUAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAUAGUGUGUCAUGUCUUUUGCAUGUGCCAC---- ((((((((((....))))))))))...((((((((((.......))))))))))...((((...((((..(((((.....)))))..))))......)))).......---- ( -24.30) >DroSim_CAF1 9093 108 + 1 CUCUAUAUAUGCAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAUAGUGUGUCGUGUCUUUUGCAUGUGCCAC---- .......(((((((((((((((((.((((((((((((.......))))))))))..))....))).))).)))))))))))(((.(((((((.....))))).)))))---- ( -26.20) >DroEre_CAF1 8369 106 + 1 CUCUAUAUGAAUACAUAUAUAGAGUUCUAGACCGAUUCACAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAUUGCUUGUAUAGU--GUCGUGUCUUUUGCAUGCGCCAC---- ...((((((((.((((...(((....)))((((((((.......)))))))).....)))).))))))))...........((--(.(((((.....))))))))...---- ( -22.70) >DroYak_CAF1 8475 108 + 1 CUCUAUAUAAAUA--UAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUACUGCUUGUAUAGU--GUCGUGUCUUUUGCAUGUGCCACCCUC (((((((((....--)))))))))...((((((((((.......)))))))))).(((.....)))(((((.....)))))((--(.(((((.....)))))...))).... ( -23.80) >consensus CUCUAUAUAUGUAGGUAUAUAGAGUUCUAGACCGAUUCAAAUUCAGUUGGUUUGUUGAUGUUUUCAUGUAAUGCUUGUAUAGU__GUCGUGUCUUUUGCAUGUGCCAC____ ((((((((((....))))))))))...((((((((((.......))))))))))...((((...((((....(((.....)))....))))......))))........... (-21.72 = -21.64 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:02 2006