
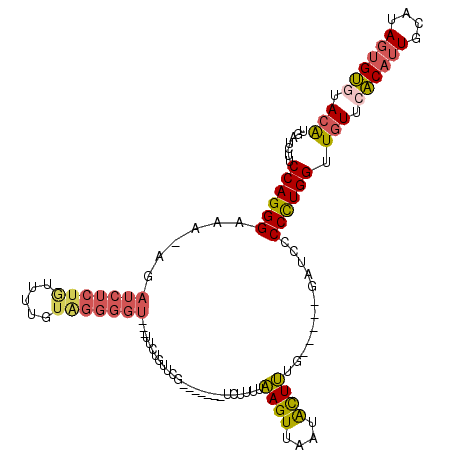
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,246,760 – 11,246,862 |
| Length | 102 |
| Max. P | 0.751126 |

| Location | 11,246,760 – 11,246,862 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11246760 102 + 22224390 UGAUCUUCCAGGGAGA-UGAUCUCUGUUUUGUAGGGGU---UUCUGUUCG-------UCUUUUAAGUUAAUACUUUG-----GAUUCCCCUGGUUGUUCACAUUGCAUAGUGUGUACA .......(((((((((-.((((((((.....)))))))---))))....(-------(((...((((....)))).)-----)))..)))))).(((.(((((......))))).))) ( -28.60) >DroSec_CAF1 6236 102 + 1 UGAUCUUCCAGGGAAA-AGAUCUCUAUUUUGUAGGGGU---UUCUGUUCG-------UCUUUUAAGUUAAUACUUUG-----GAUCCCCCUGGUUGUUCACAUUGCAUAGUGUGUACA .......((((((...-((((((((((...))))))))---))......(-------(((...((((....)))).)-----)))..)))))).(((.(((((......))))).))) ( -27.90) >DroSim_CAF1 6720 102 + 1 UGAUCUUCCAGGGAAA-GGAUCUCUGUUUUGUAGGGGU---UUCUGUUCG-------UCUUUUAAGUUAAUACUUUG-----GAUCCCCCUGGUUGUUCACAUUGCAUAGUGUGUACA .......((((((.(.-.((((((((.....)))))))---)..)....(-------(((...((((....)))).)-----)))..)))))).(((.(((((......))))).))) ( -27.40) >DroYak_CAF1 6222 102 + 1 -------CCAGGGAGA-AAAUCUCUGUUUUUUGGGGGU---UUCUGUUUUAAGUUUAUCUUUUAAGUUCAUACUUUG-----GAUCCCCCUGGUUGUUCACAUUACAGAGCGUAUACG -------(((((((((-((..(((........)))..)---))))...........((((...((((....)))).)-----)))..))))))..((((........))))....... ( -24.70) >DroAna_CAF1 11255 94 + 1 ------UCCAGGGAUACAG----------AGAAGGGAGUUAUUUCGUUAU-------CCUUUUGAGUUGAAGUUCUGUUAUUGA-GCCCUUGGUUCUGAGCAUUACAUACUGCGUACA ------.((((((...(((----------.(((((((............)-------))))))...)))..((((.......))-))))))))......(((........)))..... ( -21.00) >consensus UGAUCUUCCAGGGAAA_AGAUCUCUGUUUUGUAGGGGU___UUCUGUUCG_______UCUUUUAAGUUAAUACUUUG_____GAUCCCCCUGGUUGUUCACAUUGCAUAGUGUGUACA .......((((((......(((((((.....))))))).........................((((....))))............)))))).(((.((((((....)))))).))) (-15.18 = -16.06 + 0.88)


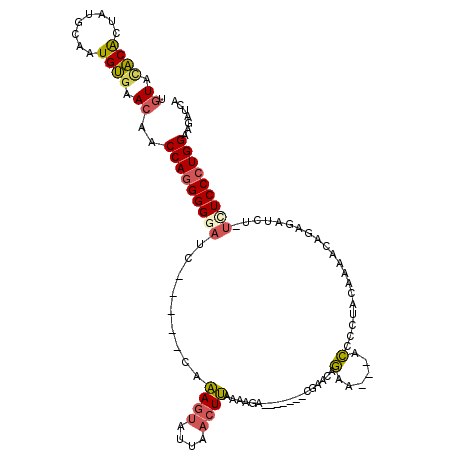
| Location | 11,246,760 – 11,246,862 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.80 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11246760 102 - 22224390 UGUACACACUAUGCAAUGUGAACAACCAGGGGAAUC-----CAAAGUAUUAACUUAAAAGA-------CGAACAGAA---ACCCCUACAAAACAGAGAUCA-UCUCCCUGGAAGAUCA (((.((((........)))).)))...(((((....-----..((((....))))......-------(.....)..---.)))))..........((((.-(((....))).)))). ( -18.30) >DroSec_CAF1 6236 102 - 1 UGUACACACUAUGCAAUGUGAACAACCAGGGGGAUC-----CAAAGUAUUAACUUAAAAGA-------CGAACAGAA---ACCCCUACAAAAUAGAGAUCU-UUUCCCUGGAAGAUCA (((.((((........)))).)))...(((((..((-----..((((....))))....(.-------....).)).---.)))))..........(((((-((......))))))). ( -20.90) >DroSim_CAF1 6720 102 - 1 UGUACACACUAUGCAAUGUGAACAACCAGGGGGAUC-----CAAAGUAUUAACUUAAAAGA-------CGAACAGAA---ACCCCUACAAAACAGAGAUCC-UUUCCCUGGAAGAUCA (((.((((........)))).))).(((((((((((-----(.((((....))))......-------.....((..---....))........).)))))-...))))))....... ( -22.20) >DroYak_CAF1 6222 102 - 1 CGUAUACGCUCUGUAAUGUGAACAACCAGGGGGAUC-----CAAAGUAUGAACUUAAAAGAUAAACUUAAAACAGAA---ACCCCCAAAAAACAGAGAUUU-UCUCCCUGG------- ........((((((..((....))....(((((...-----..((((....))))..(((.....))).........---.))))).....))))))....-.........------- ( -20.30) >DroAna_CAF1 11255 94 - 1 UGUACGCAGUAUGUAAUGCUCAGAACCAAGGGC-UCAAUAACAGAACUUCAACUCAAAAGG-------AUAACGAAAUAACUCCCUUCU----------CUGUAUCCCUGGA------ .....(((........)))......(((.(((.-...(((.((((.(((........))).-------.....(((.........))))----------)))))))))))).------ ( -11.80) >consensus UGUACACACUAUGCAAUGUGAACAACCAGGGGGAUC_____CAAAGUAUUAACUUAAAAGA_______CGAACAGAA___ACCCCUACAAAACAGAGAUCU_UCUCCCUGGAAGAUCA .((.((((........)))).))..(((((((((.........((((....))))...................(......)....................)))))))))....... (-11.52 = -11.80 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:00 2006