
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,234,056 – 11,234,198 |
| Length | 142 |
| Max. P | 0.999937 |

| Location | 11,234,056 – 11,234,163 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -23.90 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11234056 107 + 22224390 ------UUGCCCAUUUUCCUGGCUUUUUAACGAAUCGUUACUUACUUAAGACGCUUAAGGCGCUCGGCCUUCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ------..............(((....(((((...))))).........((((((((((((.....)))))(((((....)))))..)))))))))).....(((....))). ( -28.60) >DroSec_CAF1 20988 107 + 1 ------UUGCCCAUUUUCCUGCCUUUAUAACGAAUCGUUUCUUACUUAAGACGCUUAAGGCGCUCGGCCUUCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ------..............(((.....((((....((.((((....)))).))....((((((.(((((.(........).))))).)))))).........))))...))) ( -27.20) >DroSim_CAF1 12803 107 + 1 ------CUGCCCAUUUUCCUGCCUUUAUAACGAAUCGUUACUUACUUAAGACGCUUAAGGCGCUCGGCCUUCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ------..............((.....(((((...))))).........((((((((((((.....)))))(((((....)))))..)))))))))......(((....))). ( -25.00) >DroEre_CAF1 20836 113 + 1 ACCAUUUUGCCCAUUUUCCUGCCUUUUUAACGAAUCGUUACUUACUUAAGACGCUUAAGGCGCUCGGCCUCCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ....................(((.....((((...((((..........)))).....((((((.((((..(........)..)))).)))))).........))))...))) ( -25.60) >DroYak_CAF1 12858 112 + 1 GCAUUUUUACCCAUUUUCCUGCCUU-UUAACGAAUCGUUACUUACUUAAGACGCUUAAGGCGCUCGGCCUCCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ....................(((..-..((((...((((..........)))).....((((((.((((..(........)..)))).)))))).........))))...))) ( -26.20) >consensus ______UUGCCCAUUUUCCUGCCUUUUUAACGAAUCGUUACUUACUUAAGACGCUUAAGGCGCUCGGCCUUCUCGCUUUUGUGGGCCAAGCGUCGCCCAUUUCCGUUUCCGGU ....................(((.....((((...((((..........)))).....((((((.((((..(........)..)))).)))))).........))))...))) (-23.90 = -24.10 + 0.20)

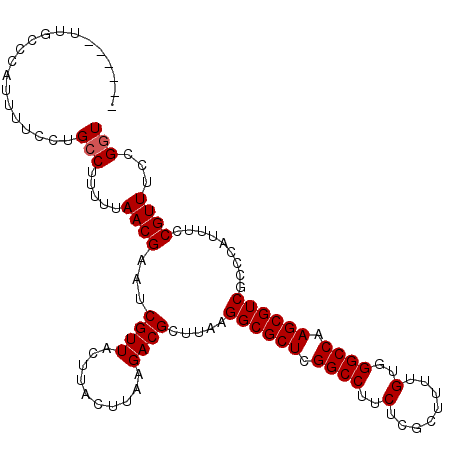
| Location | 11,234,056 – 11,234,163 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -33.71 |
| Consensus MFE | -32.18 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11234056 107 - 22224390 ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGUAACGAUUCGUUAAAAAGCCAGGAAAAUGGGCAA------ .(((....))).((((((((.(((((((((..(........)..))))))))).((((((...)))))).......)).)))))).....(((.........)))..------ ( -33.60) >DroSec_CAF1 20988 107 - 1 ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGAAACGAUUCGUUAUAAAGGCAGGAAAAUGGGCAA------ .(((....)))......((...((((((((..(........)..))))))))(((((.....((((....))))((((...))))...)))))..........))..------ ( -32.80) >DroSim_CAF1 12803 107 - 1 ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGUAACGAUUCGUUAUAAAGGCAGGAAAAUGGGCAG------ .(((....)))......((...((((((((..(........)..))))))))(((((..((.......))..((((((...)))))).)))))..........))..------ ( -34.00) >DroEre_CAF1 20836 113 - 1 ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGGAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGUAACGAUUCGUUAAAAAGGCAGGAAAAUGGGCAAAAUGGU .(((....)))...........((((((((..(........)..))))))))((((......((((.......(((((...))))).......))))....))))........ ( -34.64) >DroYak_CAF1 12858 112 - 1 ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGGAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGUAACGAUUCGUUAA-AAGGCAGGAAAAUGGGUAAAAAUGC .(((....)))......(((..((((((((..(........)..))))))))((((......((((..((...(((((...))))).-...))))))....)))).....))) ( -33.50) >consensus ACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAGUAACGAUUCGUUAAAAAGGCAGGAAAAUGGGCAA______ .(((....)))...........((((((((..(........)..))))))))((((......((((.......(((((...))))).......))))....))))........ (-32.18 = -32.22 + 0.04)


| Location | 11,234,090 – 11,234,198 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11234090 108 - 22224390 GCCAGUUUAGCUACAACGAGCAACAACAACAGCGCACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((.((((.(((......)))...............(((....))))))).)))(((((((((((..(........)..))))))))((.....)))))......... ( -35.30) >DroSec_CAF1 21022 108 - 1 GCCAGUUUAGCUACAACGAGCAACAACAACAGCGCACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((.((((.(((......)))...............(((....))))))).)))(((((((((((..(........)..))))))))((.....)))))......... ( -35.30) >DroSim_CAF1 12837 108 - 1 GCCAGUUUAGCUACAACGAGCAACAACAACAGCGCACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((.((((.(((......)))...............(((....))))))).)))(((((((((((..(........)..))))))))((.....)))))......... ( -35.30) >DroEre_CAF1 20876 105 - 1 GCCAGGUUAGCUACAACGGGCAACAACAACA---CACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGGAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((..(((.(((......)))))).......---..(((....))).....)))(((((((((((..(........)..))))))))((.....)))))......... ( -37.50) >DroYak_CAF1 12897 105 - 1 GCCAGGUUAGCUACAACGAGCAACAACAACA---CACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGGAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((..(((.(((......)))))).......---..(((....))).....)))(((((((((((..(........)..))))))))((.....)))))......... ( -37.50) >consensus GCCAGUUUAGCUACAACGAGCAACAACAACAGCGCACCGGAAACGGAAAUGGGCGACGCUUGGCCCACAAAAGCGAGAAGGCCGAGCGCCUUAAGCGUCUUAAGUAAG (((......(((......)))...............(((....))).....)))(((((((((((..(........)..))))))))((.....)))))......... (-35.26 = -35.10 + -0.16)

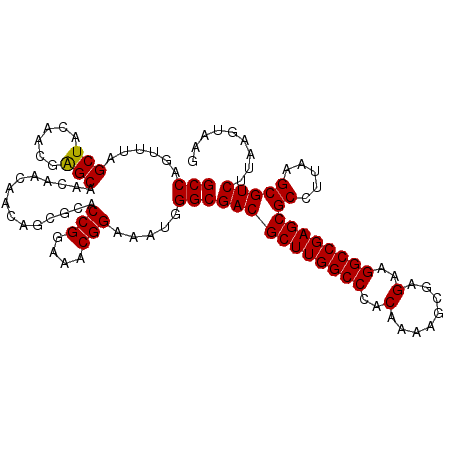
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:52 2006