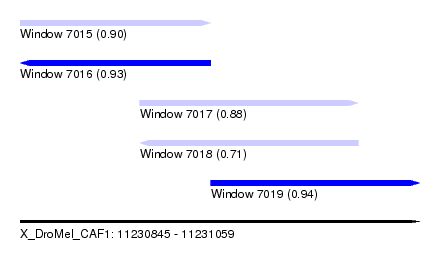
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,230,845 – 11,231,059 |
| Length | 214 |
| Max. P | 0.943761 |

| Location | 11,230,845 – 11,230,947 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 97.55 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11230845 102 + 22224390 AGGUGAACAACGGGGCGGAUGAGCAACAUCAGGCAUGCUUCCAAAUAUUACGUAUACGCCCCACGUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCAC .(((((.(...((((((((((.....)))).((.......))..............))))))...(((......)))...).)))))............... ( -27.30) >DroSim_CAF1 9609 102 + 1 AGGUGAACAACGGGGCGGAUGAGCAACAUCAGGCAUGAUUCCAAAUAUUACGUAUACGCCCCACGUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCAC .(((((.(...((((((((((.....))))..((.((((.......)))).))...))))))...(((......)))...).)))))............... ( -27.80) >DroEre_CAF1 17543 102 + 1 AGGAGAACAACGGGGCGGAUGAGCAACAUCAGGCAUGCUUCCAAAUAUUACGUAUACGCCCCACGUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCAC .((.((.....((((((((((.....)))).((.......))..............))))))...(((......))).))(((...........))).)).. ( -22.70) >DroYak_CAF1 9645 102 + 1 AGGUGAACAACGGGGCGGAUGAACGACAUCAGGCAUGCUUCCAAAUAUUACGUAUACGCCCCACGUGCCAAACAGCAAUCGCUCACCAAUUAAAAGCACCAC .(((((.(...((((((((((.....)))).((.......))..............))))))...(((......)))...).)))))............... ( -27.30) >consensus AGGUGAACAACGGGGCGGAUGAGCAACAUCAGGCAUGCUUCCAAAUAUUACGUAUACGCCCCACGUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCAC .(((((.(...((((((((((.....)))).((.......))..............))))))...(((......)))...).)))))............... (-25.98 = -26.22 + 0.25)


| Location | 11,230,845 – 11,230,947 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 97.55 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -35.41 |
| Energy contribution | -35.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11230845 102 - 22224390 GUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCACGUGGGGCGUAUACGUAAUAUUUGGAAGCAUGCCUGAUGUUGCUCAUCCGCCCCGUUGUUCACCU ...............((((((((((((((....))))...((((((.((..((((((((.((.......)).))))))))..)).)))))))))))))))). ( -38.30) >DroSim_CAF1 9609 102 - 1 GUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCACGUGGGGCGUAUACGUAAUAUUUGGAAUCAUGCCUGAUGUUGCUCAUCCGCCCCGUUGUUCACCU ...............((((((((((((((....))))...((((((.((..((((((((.((.......)).))))))))..)).)))))))))))))))). ( -38.30) >DroEre_CAF1 17543 102 - 1 GUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCACGUGGGGCGUAUACGUAAUAUUUGGAAGCAUGCCUGAUGUUGCUCAUCCGCCCCGUUGUUCUCCU ...............((.(((((((((((....))))...((((((.((..((((((((.((.......)).))))))))..)).))))))))))))).)). ( -34.30) >DroYak_CAF1 9645 102 - 1 GUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUUGGCACGUGGGGCGUAUACGUAAUAUUUGGAAGCAUGCCUGAUGUCGUUCAUCCGCCCCGUUGUUCACCU ...............((((((((((((((....))))...((((((...(((..(((((.((.......)).)))))))).....)))))))))))))))). ( -34.40) >consensus GUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCACGUGGGGCGUAUACGUAAUAUUUGGAAGCAUGCCUGAUGUUGCUCAUCCGCCCCGUUGUUCACCU ...............((((((((((((((....))))...((((((.((..((((((((.((.......)).))))))))..)).)))))))))))))))). (-35.41 = -35.73 + 0.31)


| Location | 11,230,909 – 11,231,026 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -25.91 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11230909 117 + 22224390 GUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCACCAGGUGGCGCAAAUUAAAAGUGAUUAAACCUGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUCUU ((((......((....))((((...((((..((.((((...)))).))...))))..))))........))))((((((((((((((.......))))))))))))))......... ( -28.40) >DroSim_CAF1 9673 117 + 1 GUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCACCAGGUGGCGCAAAUUAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUCGCUGGAAUUCGU (((((....(((....)))..............(((....)))))))).........(((........))).((((.((((((((((.......)))))))))).))))........ ( -25.40) >DroEre_CAF1 17607 117 + 1 GUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCACCAGGUGGCACAAAUGAAAAGUGAUUAAGCCCGCACAGCCGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUUGU (((((....(((....)))..............(((....)))))))).........(((........))).((((.((((((((((.......)))))))))).))))........ ( -25.80) >DroYak_CAF1 9709 117 + 1 GUGCCAAACAGCAAUCGCUCACCAAUUAAAAGCACCACCAGGUGGCACAAAUGAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUACUGGAAUUUGU (((((....(((....)))..............(((....)))))))).........(((........))).(((.(((((((((((.......))))))))))).)))........ ( -23.90) >consensus GUGCCCAACAGCAAUCGCUCACCAAUUAAAAGCACCACCAGGUGGCACAAAUGAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUCGU (((((....(((....)))..............(((....)))))))).........(((........))).(((((((((((((((.......)))))))))))))))........ (-25.91 = -26.23 + 0.31)

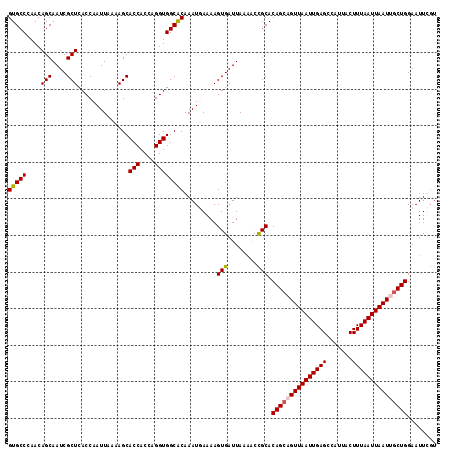
| Location | 11,230,909 – 11,231,026 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -28.49 |
| Energy contribution | -27.86 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11230909 117 - 22224390 AAGAAUUCCAGCAAUUAAUUAAAGUAAUGGCUCAAUUAACUGCUGUGCAGGUUUAAUCACUUUUAAUUUGCGCCACCUGGUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCAC ......(((((((..........(((((.(((((.....((((...)))).........(..((((...(((((((...)))))))..))))..).))))).))))))))))))... ( -35.40) >DroSim_CAF1 9673 117 - 1 ACGAAUUCCAGCGAUUAAUUAAAGUAAUGGCUCAAUUAACUGCUGUGCGGUUUUAAUCACUUUUAAUUUGCGCCACCUGGUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCAC ......((((((..........((((((.(((((...((((((...)))))).......(..((((...(((((((...)))))))..))))..).))))).))))))))))))... ( -35.80) >DroEre_CAF1 17607 117 - 1 ACAAAUUCCAGCAAUUAAUUAAAGUAAUGGCUCAAUUAACGGCUGUGCGGGCUUAAUCACUUUUCAUUUGUGCCACCUGGUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCAC ........((((((((.....((((.((((((........))))))....))))..(((((........(..((((...))))..)........)))))..))))))))........ ( -30.39) >DroYak_CAF1 9709 117 - 1 ACAAAUUCCAGUAAUUAAUUAAAGUAAUGGCUCAAUUAACUGCUGUGCGGUUUUAAUCACUUUUCAUUUGUGCCACCUGGUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUUGGCAC .(((((..(((((.((((((..(((....))).)))))).))))).((((((....(((((........(..((((...))))..)........)))))..)))))).))))).... ( -29.29) >consensus ACAAAUUCCAGCAAUUAAUUAAAGUAAUGGCUCAAUUAACUGCUGUGCGGGUUUAAUCACUUUUAAUUUGCGCCACCUGGUGGUGCUUUUAAUUGGUGAGCGAUUGCUGUUGGGCAC ........((((((((.......(((.((((..........)))))))........(((((........(((((((...)))))))........)))))..))))))))........ (-28.49 = -27.86 + -0.62)

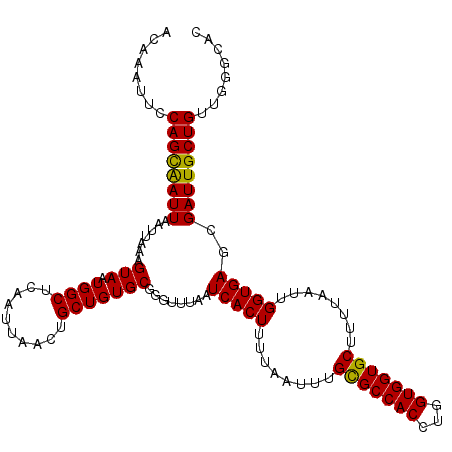
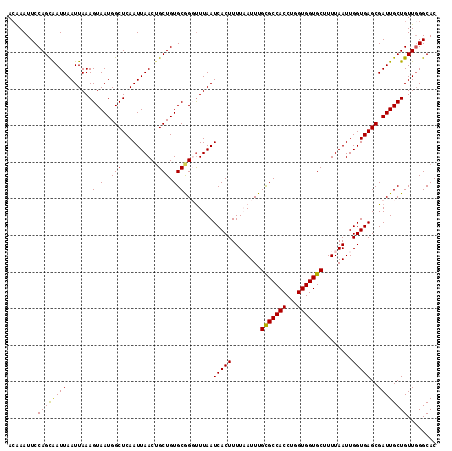
| Location | 11,230,947 – 11,231,059 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.57 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11230947 112 + 22224390 CAGGUGGCGCAAAUUAAAAGUGAUUAAACCUGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUCUUAAUAAAUUCACAGCCAUACGGUUGACACGUUUA (((((..(((.........))).....)))))..(((((((((((((((.......)))))))))))))))(((((.......))))).(((((....)))))......... ( -30.00) >DroSim_CAF1 9711 112 + 1 CAGGUGGCGCAAAUUAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUCGCUGGAAUUCGUAAUAAAUUCACAGCCAUACGGUUGACACGUUUA ...((((((..........(((........))).((((.((((((((((.......)))))))))).)))).....)))..........(((((....))))).)))..... ( -24.30) >DroEre_CAF1 17645 110 + 1 CAGGUGGCACAAAUGAAAAGUGAUUAAGCCCGCACAGCCGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUUGUGAUA--UUCACAGCCAUACGGUUGACACGUUUA ...(((.(((((((.....(((........))).((((.((((((((((.......)))))))))).))))..)))))))...--....(((((....))))).)))..... ( -29.40) >DroYak_CAF1 9747 111 + 1 CAGGUGGCACAAAUGAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUACUGGAAUUUGUGAUA-GUUCACAGCCAUACGGUUGACACAUUUU ...(((.(((((((.....(((........))).(((.(((((((((((.......))))))))))).)))..)))))))...-.....(((((....))))).)))..... ( -26.60) >consensus CAGGUGGCACAAAUGAAAAGUGAUUAAAACCGCACAGCAGUUAAUUGAGCCAUUACUUUAAUUAAUUGCUGGAAUUCGUAAUA_AUUCACAGCCAUACGGUUGACACGUUUA ...(((.(((((((.....(((........))).(((((((((((((((.......)))))))))))))))..))))))).........(((((....))))).)))..... (-24.01 = -25.57 + 1.56)

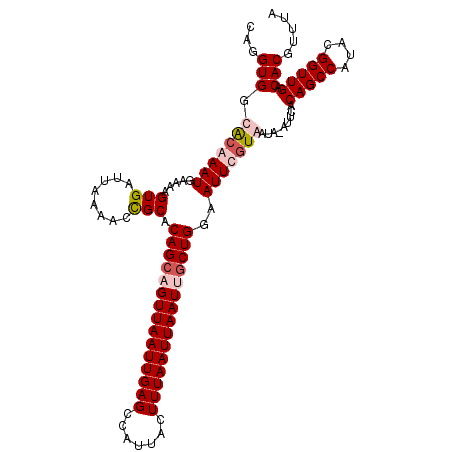

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:49 2006