| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,225,960 – 11,226,097 |
| Length | 137 |
| Max. P | 0.656479 |

| Location | 11,225,960 – 11,226,073 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.10 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -4.10 |
| Energy contribution | -4.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11225960 113 + 22224390 AACAUCUAAAUGUUAUUUAACUUAUGCAUCAAGUUGUUUAACUUGAUUUUUCCGAUCACUCGGCGGUUGGUUGUUGCAUUUUUCUCAGUGUACUUUCGCACCACAAGUUAACU (((((....)))))..(((((((..((((((((((....)))))))).....(((....)))))((((((..((.(((((......)))))))..))).)))..))))))).. ( -23.50) >DroSec_CAF1 13277 97 + 1 AACAUUUAAUUGUUUUUUAACUUAUGCAUCAAUUU---AAACU--AUUUUUCC-----------GUUUGGUUGUGGCAUUUUUCUCAGUGUACCUUCGCACUACAAGUUAACU ((((......))))..(((((((((((..(((((.---((((.--........-----------)))))))))..)))).......(((((......)))))..))))))).. ( -15.80) >DroSim_CAF1 5110 97 + 1 UACAUCUCAUUGUUAUUUAACUUAUGCAUCAAGUU---AAACU--AUUUUUCC-----------GGUUGAUUGUUGCAUUUUUCUCAGUGUAAUUUCGCACUACAAGUUAACU ................((((((((((((.(((...---.((((--........-----------))))..))).))))).......(((((......)))))..))))))).. ( -16.90) >DroEre_CAF1 12976 106 + 1 AUCAUCUAA--GUCAUAUAACUUGUGCGUUAAGUG---AAACU--AUUUUUCUAGUCAUUUGGAAGUUGGCUUCGGCAUUUUUCGAAGUGUAUUUUUGCACUACCCGUUAACU ....(((((--(((((.((((......)))).)))---).(((--(......))))..))))))(((((((.((((......))))((((((....))))))....))))))) ( -22.30) >DroYak_CAF1 4924 88 + 1 -----------GAUAUAUAACUUCUGCAUUAAAUC---CAACU--AUUUUUCUGGACAG-------UUGGCCAUUGCUUUUUUUCCAGUGUAUUUU--CACUACACGUUAACU -----------......((((....(((......(---(((((--.(((....))).))-------))))....)))..........(((((....--...)))))))))... ( -15.10) >consensus AACAUCUAA_UGUUAUUUAACUUAUGCAUCAAGUU___AAACU__AUUUUUCC___CA______GGUUGGUUGUUGCAUUUUUCUCAGUGUACUUUCGCACUACAAGUUAACU .................((((..(((((((((..................................)))))....)))).......(((((......)))))....))))... ( -4.10 = -4.34 + 0.24)


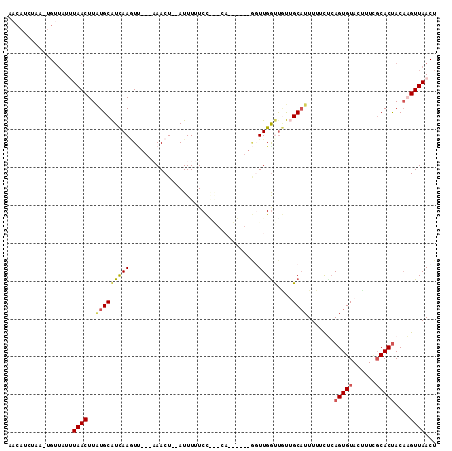
| Location | 11,225,960 – 11,226,073 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -5.42 |
| Energy contribution | -5.90 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11225960 113 - 22224390 AGUUAACUUGUGGUGCGAAAGUACACUGAGAAAAAUGCAACAACCAACCGCCGAGUGAUCGGAAAAAUCAAGUUAAACAACUUGAUGCAUAAGUUAAAUAACAUUUAGAUGUU ..((((((((((((((....))))..........................((((....))))....((((((((....)))))))).))))))))))..(((((....))))) ( -31.00) >DroSec_CAF1 13277 97 - 1 AGUUAACUUGUAGUGCGAAGGUACACUGAGAAAAAUGCCACAACCAAAC-----------GGAAAAAU--AGUUU---AAAUUGAUGCAUAAGUUAAAAAACAAUUAAAUGUU ..(((((((((.((((....))))............((..(((..((((-----------........--.))))---...)))..)))))))))))................ ( -15.40) >DroSim_CAF1 5110 97 - 1 AGUUAACUUGUAGUGCGAAAUUACACUGAGAAAAAUGCAACAAUCAACC-----------GGAAAAAU--AGUUU---AACUUGAUGCAUAAGUUAAAUAACAAUGAGAUGUA ..(((((((.(((((........)))))......(((((....((((..-----------.(((....--..)))---...))))))))))))))))................ ( -12.80) >DroEre_CAF1 12976 106 - 1 AGUUAACGGGUAGUGCAAAAAUACACUUCGAAAAAUGCCGAAGCCAACUUCCAAAUGACUAGAAAAAU--AGUUU---CACUUAACGCACAAGUUAUAUGAC--UUAGAUGAU ............((((.........(((((........))))).............(((((......)--)))).---........))))(((((....)))--))....... ( -15.90) >DroYak_CAF1 4924 88 - 1 AGUUAACGUGUAGUG--AAAAUACACUGGAAAAAAAGCAAUGGCCAA-------CUGUCCAGAAAAAU--AGUUG---GAUUUAAUGCAGAAGUUAUAUAUC----------- ...(((((((((...--....)))))..........(((.((.((((-------((((........))--)))))---)...)).)))....))))......----------- ( -18.50) >consensus AGUUAACUUGUAGUGCGAAAAUACACUGAGAAAAAUGCAACAACCAACC______UG___GGAAAAAU__AGUUU___AACUUGAUGCAUAAGUUAAAUAACA_UUAGAUGUU ...((((((((((((........)))).........(((.(((......................................))).)))))))))))................. ( -5.42 = -5.90 + 0.48)

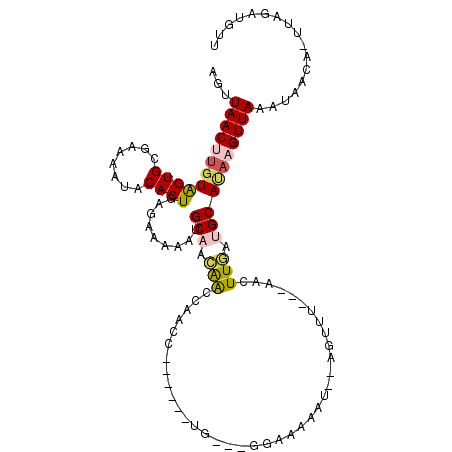

| Location | 11,226,000 – 11,226,097 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -12.96 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11226000 97 + 22224390 ACUUGAUUUUUCCGAUCACUCGGCGGUUGGUUGUUGCAUUUUUCUCAGUGUACUUUCGCACCACAAGUUAACUGCAGAAUGCCAACAAAGUUUUCGA ((((.......((((....))))..((((((..(((((.((..((..((((......))))....))..)).)))))...)))))).))))...... ( -22.00) >DroSec_CAF1 13314 84 + 1 ACU--AUUUUUCC-----------GUUUGGUUGUGGCAUUUUUCUCAGUGUACCUUCGCACUACAAGUUAACUGCAGAAUGCCAACAAAGUUUUCGA ...--.......(-----------(.....((((((((((((....(((((......)))))....((.....)))))))))).))))......)). ( -17.20) >DroSim_CAF1 5147 84 + 1 ACU--AUUUUUCC-----------GGUUGAUUGUUGCAUUUUUCUCAGUGUAAUUUCGCACUACAAGUUAACUGCAGAAUGCCAACAAAGUUUUCGA ...--.......(-----------((....((((((((((((...((((.((((((........)))))))))).)))))).)))))).....))). ( -15.20) >DroEre_CAF1 13011 95 + 1 ACU--AUUUUUCUAGUCAUUUGGAAGUUGGCUUCGGCAUUUUUCGAAGUGUAUUUUUGCACUACCCGUUAACUGCAAUAUGCCAACAGAGUUUUCGA ...--....((((((....))))))((((((....(((.((..((.((((((....))))))...))..)).))).....))))))........... ( -24.50) >DroYak_CAF1 4950 86 + 1 ACU--AUUUUUCUGGACAG-------UUGGCCAUUGCUUUUUUUCCAGUGUAUUUU--CACUACACGUUAACUGCAAUAUGCCAACAGAGCUUUCGA ...--.............(-------(((((.(((((..........(((((....--...))))).......)))))..))))))........... ( -17.73) >consensus ACU__AUUUUUCC___CA______GGUUGGUUGUUGCAUUUUUCUCAGUGUACUUUCGCACUACAAGUUAACUGCAGAAUGCCAACAAAGUUUUCGA .........................((((((..(((((.((..((.(((((......)))))...))..)).)))))...))))))........... (-12.96 = -14.20 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:45 2006