
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,184,987 – 11,185,139 |
| Length | 152 |
| Max. P | 0.969494 |

| Location | 11,184,987 – 11,185,099 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -33.08 |
| Energy contribution | -32.45 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11184987 112 + 22224390 CUAGAAACAGGUGUGAUUAGCCAAGAGAGCAUCACUGUUCUGUUCUGGGUGCCAAGAACCAAAGUCUGGCAACGACUGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGC ((((((.((((.(((((..(((....).)))))))...))))))))))(..((.(((.......)))(((((((.(((...((((.....))))..)))))))))).))..) ( -40.10) >DroSim_CAF1 4967 112 + 1 CUAGAAAAAGCGGUGAUUAGCCAAGAGAGCAUCACUAUUCUGUUCUGGCUGCCAAGAACCAAAGUCCGGCAACGACUGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGC ((((.......((((((..(((....).))))))))...(((((((((((((..........((((.(....))))))))))))))))))))))((((((...))).))).. ( -43.00) >DroEre_CAF1 4918 110 + 1 CUAGGAAA-GCUGUAAAUGGCCAUUACAGCAUCACUGUUGUGUUCUUGCUGCCAAGAACCGAAGUCUGGCAACGACCGCGGCCAGAACAGCUG-GCCGGCGUUGCCAGGCGU .(((..(.-(((((((.......))))))).)..)))(((.(((((((....)))))))))).(((((((((((....(((((((.....)))-)))).))))))))))).. ( -51.80) >DroYak_CAF1 4913 112 + 1 CUAGAAAAACCUGUAAUUUGCCAUUAGAGCAUCACUGUUCUGUUCUUGCUGCUAAGAACCGAAGUCUGGCAGCGCCCGCAGCCAGAACAGCUGGGCUGGCGUUGCCAGGUGU ................((((.....((((((....))))))(((((((....))))))))))).((((((((((((.((..((((.....)))))).))))))))))))... ( -40.40) >consensus CUAGAAAAAGCUGUAAUUAGCCAAGAGAGCAUCACUGUUCUGUUCUGGCUGCCAAGAACCAAAGUCUGGCAACGACCGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGC ...................(((...((((((....))))))(((((((....)))))))........(((((((.(((..(((((.....))).)))))))))))).))).. (-33.08 = -32.45 + -0.63)

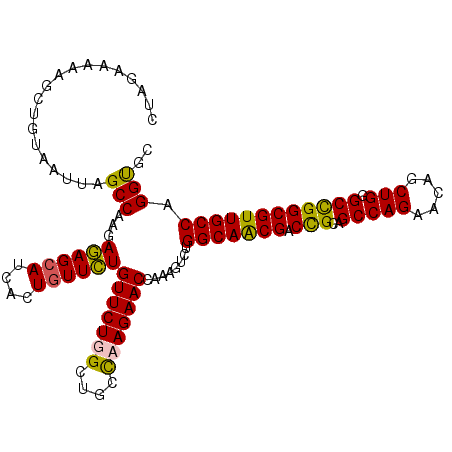
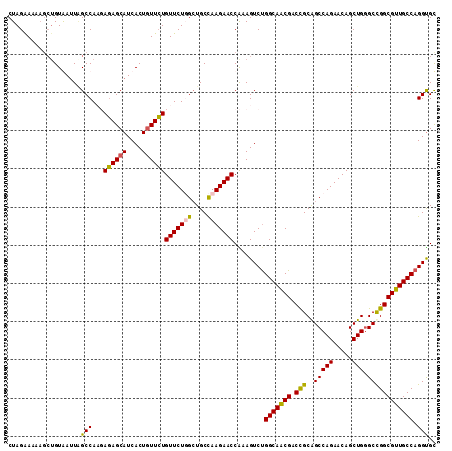
| Location | 11,184,987 – 11,185,099 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -41.02 |
| Consensus MFE | -32.51 |
| Energy contribution | -32.95 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11184987 112 - 22224390 GCACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCAGACUUUGGUUCUUGGCACCCAGAACAGAACAGUGAUGCUCUCUUGGCUAAUCACACCUGUUUCUAG ....(((((...)))))..((((((((((((.(((.......(((((...))))).....))).)))))))))....((((((((.....)))..)))))..)))....... ( -35.30) >DroSim_CAF1 4967 112 - 1 GCACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCGGACUUUGGUUCUUGGCAGCCAGAACAGAAUAGUGAUGCUCUCUUGGCUAAUCACCGCUUUUUCUAG ((..(((((...))))).....(((((((((((((.......((((.....)))).....)))))))))))))....((((((((.....)))..))))).))......... ( -40.70) >DroEre_CAF1 4918 110 - 1 ACGCCUGGCAACGCCGGC-CAGCUGUUCUGGCCGCGGUCGUUGCCAGACUUCGGUUCUUGGCAGCAAGAACACAACAGUGAUGCUGUAAUGGCCAUUUACAGC-UUUCCUAG .(((((((((((((((((-(((.....))))))..)).)))))))))......(((((((....)))))))......)))..(((((((.......)))))))-........ ( -47.50) >DroYak_CAF1 4913 112 - 1 ACACCUGGCAACGCCAGCCCAGCUGUUCUGGCUGCGGGCGCUGCCAGACUUCGGUUCUUAGCAGCAAGAACAGAACAGUGAUGCUCUAAUGGCAAAUUACAGGUUUUUCUAG ..(((((.....((((((...(((((((((((.....))(((((..(((....)))....))))).....)))))))))...)).....))))......)))))........ ( -40.60) >consensus ACACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCAGACUUCGGUUCUUGGCAGCAAGAACAGAACAGUGAUGCUCUAAUGGCUAAUCACAGCUUUUUCUAG ....((((((((((((((.(((.....)))))))..).)))))))))......(((((((....)))))))......((((((((.....)))..)))))............ (-32.51 = -32.95 + 0.44)



| Location | 11,185,025 – 11,185,139 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11185025 114 + 22224390 UCUGUUCUGGGUGCCAAGAACCAAAGUCUGGCAACGACUGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGCAAAGAACGUAGAUGAGCAACAGAAAAGCAAACGAAAUGGA ..((((((((.(((..........((((.(....)))))))).))))))))((((...))))(((((((..(((.......)))..)).))))).................... ( -34.80) >DroSim_CAF1 5005 114 + 1 UCUGUUCUGGCUGCCAAGAACCAAAGUCCGGCAACGACUGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGCAAAUAACGUAGAUGAGCAACAGAAAAGAAAACGAAAUGAA ..((((((((((((..........((((.(....)))))))))))))))))((((...))))(((((((..(((.......)))..)).))))).................... ( -39.30) >DroEre_CAF1 4955 113 + 1 UGUGUUCUUGCUGCCAAGAACCGAAGUCUGGCAACGACCGCGGCCAGAACAGCUG-GCCGGCGUUGCCAGGCGUAAAGAACGCAGAAGAGCAACAGAAAAGCAAGCGAAAUGAA (((((((((.((((...........(((((((((((....(((((((.....)))-)))).)))))))))))((.....)))))))))))).)))................... ( -47.70) >DroYak_CAF1 4951 114 + 1 UCUGUUCUUGCUGCUAAGAACCGAAGUCUGGCAGCGCCCGCAGCCAGAACAGCUGGGCUGGCGUUGCCAGGUGUAAAGAACGUAGGCGAGCAACAGAGAAGCAAACGAAAUGAA ((((((((((((...........(..((((((((((((.((..((((.....)))))).))))))))))))..)..........)))))).))))))................. ( -45.10) >consensus UCUGUUCUGGCUGCCAAGAACCAAAGUCUGGCAACGACCGCAGCCAGAACAGCUGGGCCGGCGUUGCCAGGUGCAAAGAACGUAGAUGAGCAACAGAAAAGCAAACGAAAUGAA ((((((((((((((...........(((((((((((.(((..(((((.....))).)))))))))))))))).........)))).)))).))))))................. (-32.20 = -32.58 + 0.37)



| Location | 11,185,025 – 11,185,139 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -31.59 |
| Energy contribution | -31.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11185025 114 - 22224390 UCCAUUUCGUUUGCUUUUCUGUUGCUCAUCUACGUUCUUUGCACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCAGACUUUGGUUCUUGGCACCCAGAACAGA .............((.(((((.(((.((...((((.....))..(((((((((.(....(((((.....)))))..).)))))))))......))...)))))..))))).)). ( -32.70) >DroSim_CAF1 5005 114 - 1 UUCAUUUCGUUUUCUUUUCUGUUGCUCAUCUACGUUAUUUGCACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCGGACUUUGGUUCUUGGCAGCCAGAACAGA ..................(((.(((...............))).(((((...)))))..)))(((((((((((((.......((((.....)))).....))))))))))))). ( -35.46) >DroEre_CAF1 4955 113 - 1 UUCAUUUCGCUUGCUUUUCUGUUGCUCUUCUGCGUUCUUUACGCCUGGCAACGCCGGC-CAGCUGUUCUGGCCGCGGUCGUUGCCAGACUUCGGUUCUUGGCAGCAAGAACACA ................((((((((((.....((((.....))))((((((((((((((-(((.....))))))..)).)))))))))((....))....)))))).)))).... ( -43.90) >DroYak_CAF1 4951 114 - 1 UUCAUUUCGUUUGCUUCUCUGUUGCUCGCCUACGUUCUUUACACCUGGCAACGCCAGCCCAGCUGUUCUGGCUGCGGGCGCUGCCAGACUUCGGUUCUUAGCAGCAAGAACAGA .........((((.((((.(((((((.(((...((.....))..((((((.((((....(((((.....)))))..)))).)))))).....)))....))))))))))))))) ( -40.10) >consensus UUCAUUUCGUUUGCUUUUCUGUUGCUCAUCUACGUUCUUUACACCUGGCAACGCCGGCCCAGCUGUUCUGGCUGCAGUCGUUGCCAGACUUCGGUUCUUGGCAGCAAGAACAGA .............((.((((((((((.....((((.....))..((((((((((((((.(((.....)))))))..).)))))))))......))....)))))).)))).)). (-31.59 = -31.40 + -0.19)

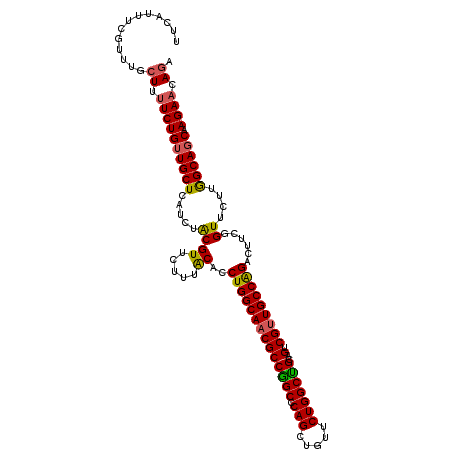
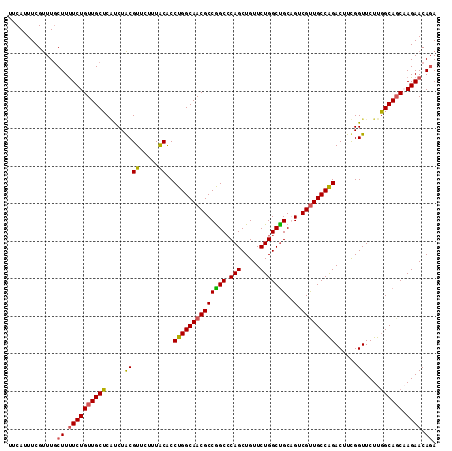
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:30 2006