| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,170,713 – 11,170,815 |
| Length | 102 |
| Max. P | 0.936267 |

| Location | 11,170,713 – 11,170,815 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
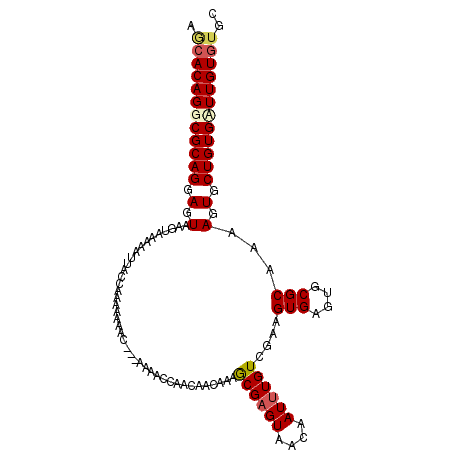
>X_DroMel_CAF1 11170713 102 + 22224390 AGCACAGGCGCAGGAGUAAGUAAAAAUUAACAUAAAAAC--AAAAACCAACAACAAAGCGAGUAACAAUUUGUCGAAGUGAGUGCGCAAAAGUGCUGUGCUUGU---- ...(((((((((...........................--.........((.....((((((....)))))).....))...((((....)))))))))))))---- ( -19.40) >DroSec_CAF1 5324 108 + 1 AGCACAGGCGCAGGAGUAAGUAAAAAUUACAAAAAAAAAAAAAAAACCAACAACAAAGCGAGUAAUAAUUUGUCGAAGUGAGUGCGCAAAAGUGCUGUGAUUGUGUGC (((((..(((((...((((.......)))).......................((...(((.(((....))))))...))..)))))....)))))............ ( -19.80) >DroSim_CAF1 833 102 + 1 AACACAGGCGCAGGAGUAAGUAAAAAUUACCAAAAAAAUAA------CAACAACAAAGCGAGUAAUAAUUUGUCGAAGUGAGUGCGCAUAAGUGCUGUGAUUGUGUGC .((((((.((((.(.((((.......)))))..........------......((...(((.(((....))))))...))...((((....)))))))).)))))).. ( -18.20) >DroYak_CAF1 832 97 + 1 AGCACAGGCGCAGGAGUAAGUAAAAAAAUU-AAAAACAC---GAAAACA-------UACGAGUAACAAUAUGUUGAAGUGAGUGCGCAAAAGUGCUGUGUUUGUGUGC .(((((((((((..................-.....(((---...((((-------((..........))))))...)))...((((....))))))))))))))).. ( -25.80) >consensus AGCACAGGCGCAGGAGUAAGUAAAAAUUACCAAAAAAAC___AAAACCAACAACAAAGCGAGUAACAAUUUGUCGAAGUGAGUGCGCAAAAGUGCUGUGAUUGUGUGC .((((((((((((.(.(........................................((((((....))))))....(((....)))...).).)))))))))))).. (-18.10 = -18.98 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:16 2006