| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,240,172 – 1,240,271 |
| Length | 99 |
| Max. P | 0.782152 |

| Location | 1,240,172 – 1,240,271 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.23 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
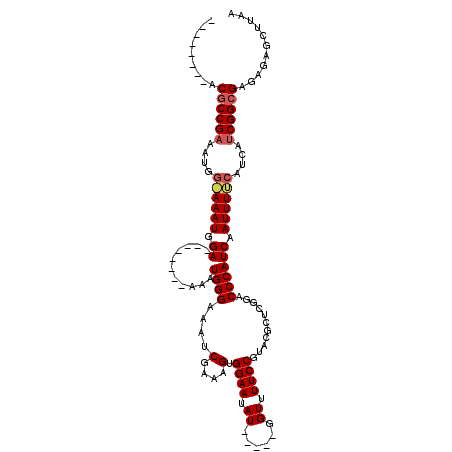
>X_DroMel_CAF1 1240172 99 + 22224390 CGGGCGGAACGCCG-------AAAUGGA--------AAAUGGGAAAUCGAAAGUGGAAUAU-----GGUUUUCCGUGCGCUCGCACCCAUCAAUUUUCAUCAGCGGCGAGAGAGCUUAA .((((....(((((-------..(((((--------((..(((((((((...........)-----))))))))((((....)))).......)))))))...))))).....)))).. ( -30.40) >DroEre_CAF1 7723 98 + 1 --------ACGCCGAAAUGGGAAAUGGA--------AAAUGGGAAAUCGAAAGUGGAAUAU-----GGUUUUCCGUACGCGCGGACCCAUCAAUUUCCAUCAUCGGCGAGAGAGCUUAA --------.((((((.(((((((.((..--------..(((((...(((...(((...(((-----((....)))))))).))).))))))).)))))))..))))))........... ( -31.70) >DroYak_CAF1 7733 111 + 1 --------ACACCGAAAUGGAAAAUGGAAAAACGGAAAAUGGGAAAUCGAAAGUGGAAUAUGGAAUGGUUUUCCGUACGCUCGGACCCAUCAAUUUUCAUCAUCGGCGAGAGAGCUUAA --------.(.((((.(((((((.((......))....(((((...((((..(((...(((((((.....)))))))))))))).)))))...)))))))..)))).)........... ( -25.00) >consensus ________ACGCCGAAAUGG_AAAUGGA________AAAUGGGAAAUCGAAAGUGGAAUAU_____GGUUUUCCGUACGCUCGGACCCAUCAAUUUUCAUCAUCGGCGAGAGAGCUUAA .........((((((....((((((.((...........((((....(....).((((.((......)).))))...........)))))).))))))....))))))........... (-19.12 = -20.23 + 1.11)

| Location | 1,240,172 – 1,240,271 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -19.29 |
| Energy contribution | -20.52 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
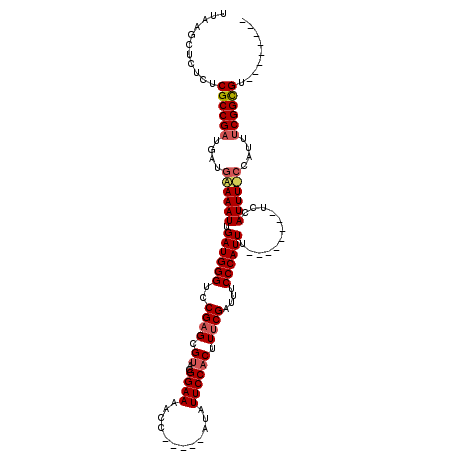
>X_DroMel_CAF1 1240172 99 - 22224390 UUAAGCUCUCUCGCCGCUGAUGAAAAUUGAUGGGUGCGAGCGCACGGAAAACC-----AUAUUCCACUUUCGAUUUCCCAUUU--------UCCAUUU-------CGGCGUUCCGCCCG ....((.....(((((..((((......(((((((((....))))((((....-----...))))............))))).--------..)))).-------)))))....))... ( -25.80) >DroEre_CAF1 7723 98 - 1 UUAAGCUCUCUCGCCGAUGAUGGAAAUUGAUGGGUCCGCGCGUACGGAAAACC-----AUAUUCCACUUUCGAUUUCCCAUUU--------UCCAUUUCCCAUUUCGGCGU-------- ...........((((((....((((((.((((((....((.....((((....-----...)))).....))....)))))).--------...))))))....)))))).-------- ( -28.40) >DroYak_CAF1 7733 111 - 1 UUAAGCUCUCUCGCCGAUGAUGAAAAUUGAUGGGUCCGAGCGUACGGAAAACCAUUCCAUAUUCCACUUUCGAUUUCCCAUUUUCCGUUUUUCCAUUUUCCAUUUCGGUGU-------- ...........((((((....((((((.((((((..((((.((..((((............)))))).))))....))))))............))))))....)))))).-------- ( -22.32) >consensus UUAAGCUCUCUCGCCGAUGAUGAAAAUUGAUGGGUCCGAGCGUACGGAAAACC_____AUAUUCCACUUUCGAUUUCCCAUUU________UCCAUUU_CCAUUUCGGCGU________ ...........((((((....((((((.((((((..((((.((..((((............)))))).))))....))))))............))))))....))))))......... (-19.29 = -20.52 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:17 2006