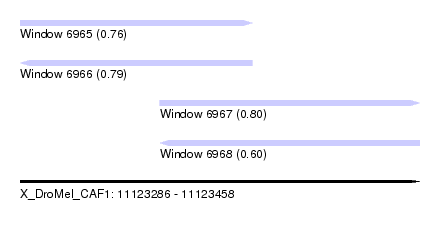
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,123,286 – 11,123,458 |
| Length | 172 |
| Max. P | 0.797206 |

| Location | 11,123,286 – 11,123,386 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11123286 100 + 22224390 UUAACGAAGCACUCGCCCCGUGGCCAAAACCCAAUUAAAAUGCAGUUAACCGAGAUUGCCUGCCUUGAUUUGGCCUGAUUAUGUUUCUGGACACUGGCUA ..............(((..(((.(((((((..(((((....((((((......))))))..(((.......))).)))))..)))).))).))).))).. ( -22.70) >DroSec_CAF1 38592 100 + 1 UUGACGAAGCACUCGCCCCGUGGCCGAAACCCAAUUAAAAUGCAGUUAACUGAGAUUGACUGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUA ..............(((..(((((((((((((((....((.((((((((......)))))))).))...)))).........))))).)))))).))).. ( -32.00) >DroEre_CAF1 37333 100 + 1 CUAACGAAACACUCGUCCAGUGGCCAAAACCCAAUUAGUAUGCAGUUAACUGGGAUUUGCUGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCUACUGGCUC ...((((.....))))((((((((((((((..((((((..(.(((....))).).......(((.......)))))))))..)))).))))))))))... ( -31.30) >DroYak_CAF1 41395 99 + 1 UCAACGAAGCACUCGCCC-GUGGCCAAAGCCCAAUUAGAAUGCAGUUAACUGAGAUUCCCAGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUA ..............(((.-(((((((((((..((((((....(((....))).........(((.......)))))))))..)))).))))))).))).. ( -32.00) >consensus UUAACGAAGCACUCGCCCCGUGGCCAAAACCCAAUUAAAAUGCAGUUAACUGAGAUUGCCUGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUA ..............(((..(((((((((((..((((((....(((....))).........(((.......)))))))))..)))).))))))).))).. (-22.40 = -22.65 + 0.25)

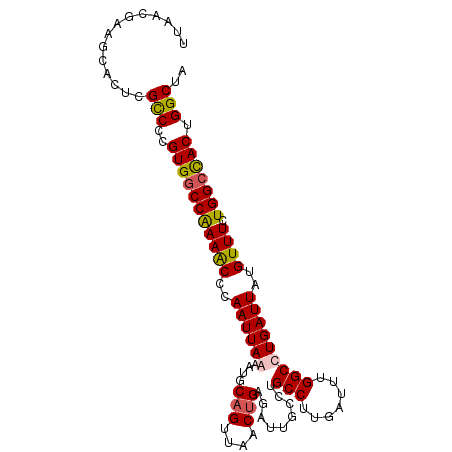

| Location | 11,123,286 – 11,123,386 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -25.41 |
| Energy contribution | -26.35 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11123286 100 - 22224390 UAGCCAGUGUCCAGAAACAUAAUCAGGCCAAAUCAAGGCAGGCAAUCUCGGUUAACUGCAUUUUAAUUGGGUUUUGGCCACGGGGCGAGUGCUUCGUUAA ......((((......)))).....(((((((((((.((((..(((....)))..)))).......))))..)))))))(((((((....)))))))... ( -27.10) >DroSec_CAF1 38592 100 - 1 UAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCAGUCAAUCUCAGUUAACUGCAUUUUAAUUGGGUUUCGGCCACGGGGCGAGUGCUUCGUCAA ...((.((((((.(((((........(((.......)))........((((((((.......)))))))))))))))))))))((((((...)))))).. ( -32.10) >DroEre_CAF1 37333 100 - 1 GAGCCAGUAGCCAGAAACAUAAUCAGGCCAAAUCAAGGCAGCAAAUCCCAGUUAACUGCAUACUAAUUGGGUUUUGGCCACUGGACGAGUGUUUCGUUAG ...(((((.(((((((..........(((.......))).......((((((((.........))))))))))))))).)))))(((((...)))))... ( -30.50) >DroYak_CAF1 41395 99 - 1 UAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCUGGGAAUCUCAGUUAACUGCAUUCUAAUUGGGCUUUGGCCAC-GGGCGAGUGCUUCGUUGA ..(((.(((((((((..(.......((((.......)))).(((((..(((....))).))))).....)..)))))))))-.))).............. ( -33.10) >consensus UAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCAGGCAAUCUCAGUUAACUGCAUUCUAAUUGGGUUUUGGCCACGGGGCGAGUGCUUCGUUAA ...((.((((((((((..........(((.......))).......(((((((((.......))))))))))))))))))).))(((((...)))))... (-25.41 = -26.35 + 0.94)

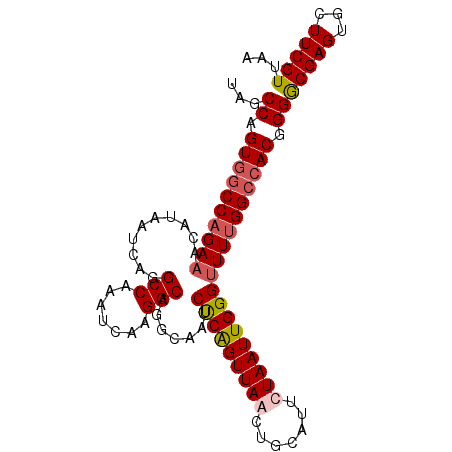
| Location | 11,123,346 – 11,123,458 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -22.41 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11123346 112 + 22224390 UGCCUUGAUUUGGCCUGAUUAUGUUUCUGGACACUGGCUAAAAUAAGAGGGCGAUAUAAUUGAAGAAAACUGAAUUUUCUCUAAUUAUA--GGAGUUCAUUA------UAAGCCAAACGA .((((((.(((((((......((((....))))..))))))).)))).((((..((((((((.((((((.....)))))).))))))))--...))))....------...))....... ( -25.40) >DroSec_CAF1 38652 114 + 1 UGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUAAAAUAAGAGGGCGAUUUAAUUGGAGGAAACUGAACUUGCUCUAAUUAUAGAGGAGUUCAUUA------UAAGCCAAAUGU .(((((....(((((.((.......)).)))))((..........))))))).......((((((....))((((((.(((((....)))))))))))....------....)))).... ( -32.20) >DroEre_CAF1 37393 112 + 1 UGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCUACUGGCUCAAAUAAGAGGGCGAUAUAAUUGGAGGAAACUGAAUUUUCUCUAAUUAUA--GGAGUUCAUUA------UAAGCCAAAUGA ..........(((((.((.......)).))))).(((((...((((..((((..(((((((((((((((.....)))))))))))))))--...)))).)))------).)))))..... ( -32.90) >DroYak_CAF1 41454 118 + 1 AGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUAAAAUAGGGUGGCGAUAUAAUUGGAGGAAACAGAAUUUUACCUAAUUAUA--GGAGUUCAUUACGAGCAUAAGCCAAAUGG .......((((((((..((((((((....((((((..(((...)))))))))))))))))..).(....).........((((....))--)).((((.....))))....))))))).. ( -31.80) >consensus UGCCUUGAUUUGGCCUGAUUAUGUUUCUGGCCACUGGCUAAAAUAAGAGGGCGAUAUAAUUGGAGGAAACUGAAUUUUCUCUAAUUAUA__GGAGUUCAUUA______UAAGCCAAAUGA .(((......(((((.((.......)).)))))..)))...........(((..(((((((((((((((.....)))))))))))))))...(....).............)))...... (-22.41 = -23.85 + 1.44)


| Location | 11,123,346 – 11,123,458 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11123346 112 - 22224390 UCGUUUGGCUUA------UAAUGAACUCC--UAUAAUUAGAGAAAAUUCAGUUUUCUUCAAUUAUAUCGCCCUCUUAUUUUAGCCAGUGUCCAGAAACAUAAUCAGGCCAAAUCAAGGCA .....(((((.(------(((.((.....--(((((((..(((((((...)))))))..)))))))......))))))...)))))((((......))))......(((.......))). ( -22.20) >DroSec_CAF1 38652 114 - 1 ACAUUUGGCUUA------UAAUGAACUCCUCUAUAAUUAGAGCAAGUUCAGUUUCCUCCAAUUAAAUCGCCCUCUUAUUUUAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCA ..(((((((((.------...((((((.(((((....)))))..))))))(((((..(((.(((((............)))))....)))...)))))......)))))))))....... ( -26.80) >DroEre_CAF1 37393 112 - 1 UCAUUUGGCUUA------UAAUGAACUCC--UAUAAUUAGAGAAAAUUCAGUUUCCUCCAAUUAUAUCGCCCUCUUAUUUGAGCCAGUAGCCAGAAACAUAAUCAGGCCAAAUCAAGGCA ...(((((((..------...((......--(((((((.(((.((((...)))).))).))))))).....(((......))).))..)))))))...........(((.......))). ( -20.90) >DroYak_CAF1 41454 118 - 1 CCAUUUGGCUUAUGCUCGUAAUGAACUCC--UAUAAUUAGGUAAAAUUCUGUUUCCUCCAAUUAUAUCGCCACCCUAUUUUAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCU (((((((((((...........(((..((--((....)))).....)))((((((.............(((((.(((...)))...)))))..)))))).....)))))))))...)).. ( -21.56) >consensus UCAUUUGGCUUA______UAAUGAACUCC__UAUAAUUAGAGAAAAUUCAGUUUCCUCCAAUUAUAUCGCCCUCUUAUUUUAGCCAGUGGCCAGAAACAUAAUCAGGCCAAAUCAAGGCA ...(((((((.........(((((.......(((((((.(((.(((.....))).))).)))))))........))))).........)))))))...........(((.......))). (-15.76 = -16.00 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:00 2006