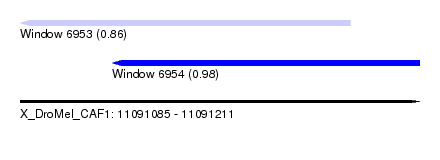
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,091,085 – 11,091,211 |
| Length | 126 |
| Max. P | 0.982944 |

| Location | 11,091,085 – 11,091,189 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.01 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
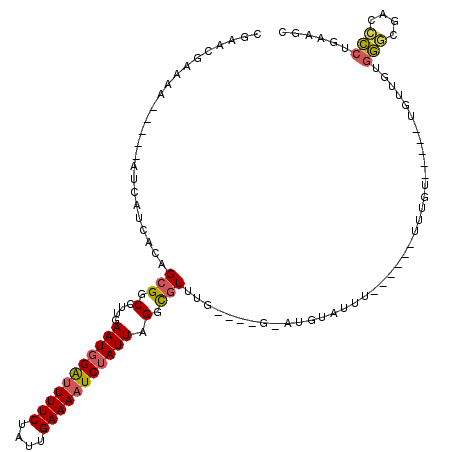
>X_DroMel_CAF1 11091085 104 - 22224390 CGAACGAAAA-----AUCAUCAGAGCGUCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGCGGUGUAUGUAUUU------UUUGU-----UGUUGUGGGCGACGCCUGAAGC ..((((((((-----((((((.(((((.(((..(((((((((((....)))))))))))))))))))..)))).....))))------)))))-----)(((.(((((...))))).))) ( -32.50) >DroSec_CAF1 10767 86 - 1 C---GGAUAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGU---------------UU------UUUGU-----UGUUGUGGGCGAGCCCUGAAGC .---((....-----.((((.((((((((((..(((((((((((....)))))))))))))))..---------------..------..)))-----))).)))).....))....... ( -25.70) >DroSim_CAF1 5109 87 - 1 CUAACGAAAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAG-CGU----------------U------UUUGU-----UGUUGAGGGCGAGCCCUGAAGC ..........-----.......(((.(((....(((((((((((....)))))))))))..-(((----------------(------(((..-----....))))))).)))))).... ( -23.50) >DroEre_CAF1 8654 104 - 1 UGGACGAA-A-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGUGGUGGCUGUUUUU-U----UACGU-----UGUUGUGGGUGACCCCUGGAGA ..((((((-(-----((((((((((..((((..(((((((((((....)))))))))))))))..)))))))))......))-)----).)))-----).....((....))........ ( -32.80) >DroYak_CAF1 11062 111 - 1 CGCACGAAAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGUCGUUUGUGUUGAAUGUAUUUUU----UUUGUUUGCCUGUUGUGGGUGACCCCUGAAGA .(((((((((-----((((.(((((((((....(((((((((((....))))))))))).)))).))))).))).......)))----)))))..))((...(.((....)).)...)). ( -32.11) >DroAna_CAF1 10912 111 - 1 CCAGCGGUUCCCGCUGCGGGCGGAGUGGCCUUGAAUAGGCUUUCUAUUGAAAAUCCAUUAGUUCUAAGG---GAAUUCAUAUUUCUCUUCUGUUU-----UUUUAAAGGGGUUAUG-AGC ...((.((((((((.....))(((...((((.....))))((((....)))).)))...........))---))))(((((..((((((..(...-----..)..)))))).))))-))) ( -27.80) >consensus CGAACGAAAA_____AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUG____G_AUGUAUUU______UUUGU_____UGUUGUGGGCGACCCCUGAAGC ........................(((.(....(((((((((((....))))))))))).).))).......................................(((....)))...... (-12.01 = -12.07 + 0.06)


| Location | 11,091,114 – 11,091,211 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.09 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.45 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11091114 97 - 22224390 UGGCGAACUUUUU-----UUUU----------UUUUG---CGAACGAAAA-----AUCAUCAGAGCGUCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGCGGUGUAUGUAU ..(((........-----..((----------(((((---....))))))-----).((((.(((((.(((..(((((((((((....)))))))))))))))))))..))))..))).. ( -25.00) >DroSec_CAF1 10796 82 - 1 UGGAGAACUUUUU-----UUUU----------UUUUGGGGC---GGAUAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGU--------------- .............-----....----------.......((---.(((..-----...)))...)).((((..(((((((((((....)))))))))))))))..--------------- ( -19.60) >DroSim_CAF1 5137 84 - 1 UGGCGAACUUUUU-----UUUU----------UUUUUUUGCUAACGAAAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAG-CGU--------------- (((((((......-----....----------....))))))).......-----............((....(((((((((((....))))))))))).)-)..--------------- ( -16.04) >DroEre_CAF1 8684 94 - 1 UGGCGAACUUC-------UUUU----------UUUUG---UGGACGAA-A-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGUGGUGGCUGUUU .(((.......-------...(----------(((((---....))))-)-----).((((((((..((((..(((((((((((....)))))))))))))))..))))))))))).... ( -31.40) >DroYak_CAF1 11098 102 - 1 UGGCGAACUUUUUCUUUUUUUU----------UUUUG---CGCACGAAAA-----AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGUCGUUUGUGUUGAAUGUAU ..(((...............((----------(((((---....))))))-----)(((.(((((((((....(((((((((((....))))))))))).)))).))))).))).))).. ( -26.00) >DroAna_CAF1 10946 112 - 1 UGGCACAUCCUUU-----UUUCCGCCCAGCAAUUUUUUGGCCAGCGGUUCCCGCUGCGGGCGGAGUGGCCUUGAAUAGGCUUUCUAUUGAAAAUCCAUUAGUUCUAAGG---GAAUUCAU .......((((((-----.((((((((..(((....)))((.(((((...))))))))))))))).(((((.....)))))..(((.((......)).)))....))))---))...... ( -37.00) >consensus UGGCGAACUUUUU_____UUUU__________UUUUG___CGAACGAAAA_____AUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUG____G_AUGUAU ................................................................(((.(....(((((((((((....))))))))))).).)))............... (-10.23 = -10.45 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:48 2006