
| Sequence ID | X_DroMel_CAF1 |
|---|---|
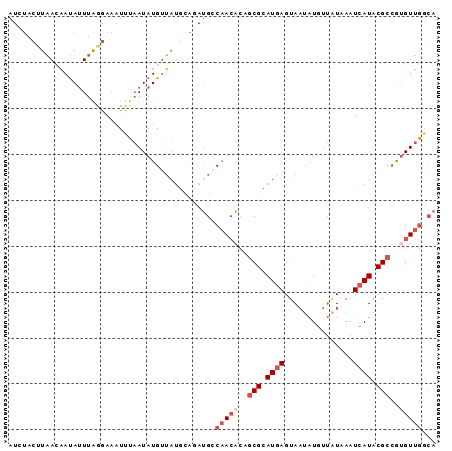
| Location | 11,090,804 – 11,090,897 |
| Length | 93 |
| Max. P | 0.999513 |

| Location | 11,090,804 – 11,090,897 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 74.09 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -5.22 |
| Energy contribution | -6.62 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11090804 93 + 22224390 AUCUACUUAACAAUAUUUAGGAAAUUUAAUAUGUUAUGCUGAUGGCAACACGGCGCAUGAGUAAUAUGUUAUAAAUCAUACGCCGCGUUGGCA (((..(.(((((.((((..........))))))))).)..)))(.((((.(((((.((((((((....))))...)))).))))).)))).). ( -23.50) >DroSec_CAF1 10527 93 + 1 AUCUACUUAACAAUAUUUAGGAAAUUUAAUAUGUUAUGCGAAUGCCAACACUGCGCAUGAGUAAUAUGUUAUAAAUCAUACGCCGUGUUGGCA .......(((((.((((..........)))))))))......(((((((((.(((.((((((((....))))...)))).))).))))))))) ( -24.10) >DroSim_CAF1 4905 76 + 1 AUCGGA-UUACUACAUUUAGAA--U--UAUAUGUA---UCGAUGCCAACA-UGCG-AUGAGUAUAGU----UAAAUCAUACGC-GUGU--GCA (((((.-....(((((......--.--...)))))---)))))((..(((-((((-((((.......----....)))).)))-))))--)). ( -17.00) >DroEre_CAF1 8369 93 + 1 AUGUGCUUAAAAAUAGUUGGGAAAUUCAAUAUGUUAUGCAGAAGACAAAUCAACGCAUAAGUAAUAUGUUAUUAAUCAUACGCCAUGUUGAAC .....(((((......)))))...(((((((((((((((.(..((....))..)))))))....((((........))))...))))))))). ( -14.20) >DroYak_CAF1 10787 93 + 1 AUGUACUUAAAAAUAUUUAAGAAAUUUAAUAUGCUAUCCAUAUGGCAACACAGCGCAUGAGUAAUAUGUUGUUAAUCAUACGCCAUGUUGACA .....((((((....)))))).......(((((.....)))))..(((((..(((.((((.((((.....)))).)))).)))..)))))... ( -17.60) >consensus AUCUACUUAACAAUAUUUAGGAAAUUUAAUAUGUUAUGCAGAUGCCAACACAGCGCAUGAGUAAUAUGUUAUAAAUCAUACGCCGUGUUGGCA .............................................(((((..(((.((((...............)))).)))..)))))... ( -5.22 = -6.62 + 1.40)


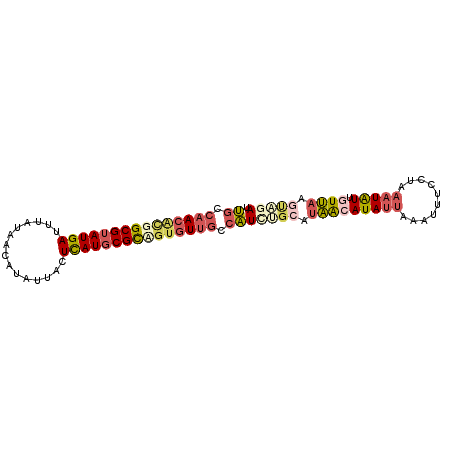
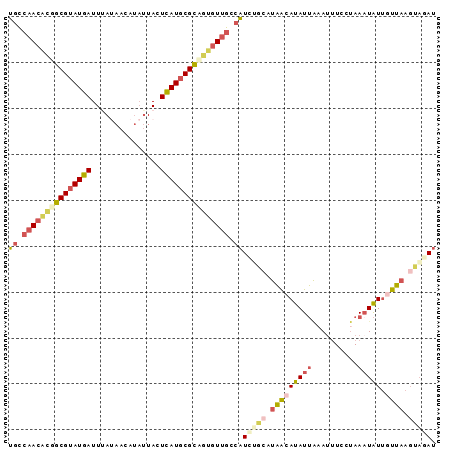
| Location | 11,090,804 – 11,090,897 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.09 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -15.88 |
| Energy contribution | -18.08 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11090804 93 - 22224390 UGCCAACGCGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCCGUGUUGCCAUCAGCAUAACAUAUUAAAUUUCCUAAAUAUUGUUAAGUAGAU ((.(((((((((((((((...(((...)))...))))))))))))))).))((.((.(((((((((..........)))).))))).)).)). ( -29.20) >DroSec_CAF1 10527 93 - 1 UGCCAACACGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCAGUGUUGGCAUUCGCAUAACAUAUUAAAUUUCCUAAAUAUUGUUAAGUAGAU (((((((((.((((((((...(((...)))...)))))))).)))))))))((..(.(((((((((..........)))).))))).)..)). ( -29.60) >DroSim_CAF1 4905 76 - 1 UGC--ACAC-GCGUAUGAUUUA----ACUAUACUCAU-CGCA-UGUUGGCAUCGA---UACAUAUA--A--UUCUAAAUGUAGUAA-UCCGAU ...--....-(((.((((....----.......))))-))).-.(((((.((...---(((((.((--.--...)).)))))...)-)))))) ( -11.40) >DroEre_CAF1 8369 93 - 1 GUUCAACAUGGCGUAUGAUUAAUAACAUAUUACUUAUGCGUUGAUUUGUCUUCUGCAUAACAUAUUGAAUUUCCCAACUAUUUUUAAGCACAU ((((((.(((((((((((.(((((...))))).)))))))).....(((.....)))...))).))))))....................... ( -13.80) >DroYak_CAF1 10787 93 - 1 UGUCAACAUGGCGUAUGAUUAACAACAUAUUACUCAUGCGCUGUGUUGCCAUAUGGAUAGCAUAUUAAAUUUCUUAAAUAUUUUUAAGUACAU .(.(((((((((((((((.(((.......))).))))))))))))))).)(((((.....))))).......((((((....))))))..... ( -25.00) >consensus UGCCAACACGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCAGUGUUGCCAUCUGCAUAACAUAUUAAAUUUCCUAAAUAUUGUUAAGUAGAU ((.(((((((((((((((...............))))))))))))))).))(((((.(((((((((..........))))).)))).))))). (-15.88 = -18.08 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:46 2006