| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,090,209 – 11,090,307 |
| Length | 98 |
| Max. P | 0.887685 |

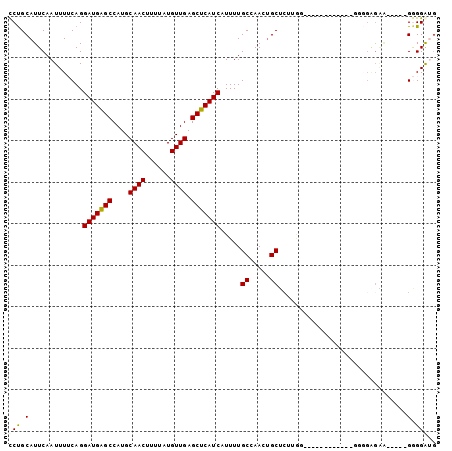
| Location | 11,090,209 – 11,090,307 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -17.18 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11090209 98 + 22224390 CCUGCAUUCAAUUUUCAGGAUGAGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCCAACUGCUCUUGUACGAUGGCAAAAAAAGGGGU-----GGGGGUG (((.(((((.........(((((((....((((......)))).))))))).((((((((.((((....))).).))))))))....))))-----).))).. ( -35.70) >DroPse_CAF1 9887 91 + 1 UCUGCAUUCAAUUUACAGGAUGGGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCAAAAUGCUCAAAG------------GGGGAGAACACCGGAGGAUG ..((((............(((((((....((((......)))).)))))))....))))..((.(((...(------------((......).)).))).)). ( -21.69) >DroSec_CAF1 9950 86 + 1 CCUGCAUUCAAUUUUCAGGAUGAGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCCAACUGCUCUUGA------------GGGGAGGA-----GGGGAGG (((.(.............(((((((....((((......)))).)))))))......((..((.(((....------------))).))..-----))).))) ( -26.20) >DroEre_CAF1 7772 89 + 1 CCUGCAUUCAAUUUUCAGGAUGAGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCCAACUGCUCUUGUGCGACGGAG---------AA-----GGGGGUG ....(((((..(((((..(((((((....((((......)))).)))))))...(((((((......))).))))..)))---------))-----..))))) ( -26.60) >DroAna_CAF1 9000 78 + 1 CCUGCAUUCAAUUUUCAGGAUGAGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCCAACGGCG--------------------GGGUG-----GGGGAGC (((.(((((.........(((((((....((((......)))).)))))))....((((...))))--------------------)))))-----.)))... ( -27.80) >DroPer_CAF1 9907 91 + 1 UCCGCAUUCAAUUUACAGGAUGGGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCAAAAUGCUCAAAG------------GGGGAGAAUACCGGAGGAUG ((((((((..........(((((((....((((......)))).))))))).........)))))......------------((........)))))..... ( -23.11) >consensus CCUGCAUUCAAUUUUCAGGAUGAGCCAUGCAACUUUUAUGUUGAGCUCAUCAUUUUGCCAACUGCUCUUGG____________GGGGAGAA_____GGGGAUG (((.(.............(((((((....((((......)))).))))))).....((.....))...............................).))).. (-17.18 = -16.93 + -0.25)


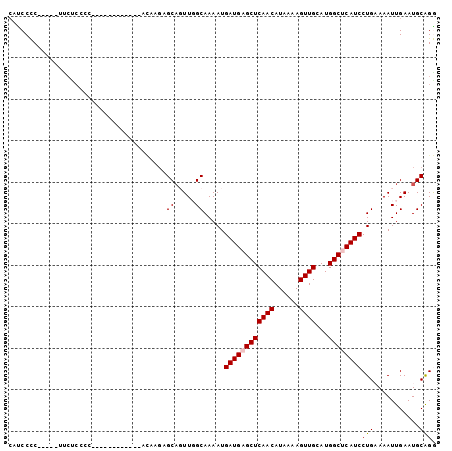
| Location | 11,090,209 – 11,090,307 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11090209 98 - 22224390 CACCCCC-----ACCCCUUUUUUUGCCAUCGUACAAGAGCAGUUGGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCUCAUCCUGAAAAUUGAAUGCAGG .......-----...(((..((((((((..((......))...)))))))).((((((((((((......))))...))))))))...............))) ( -24.20) >DroPse_CAF1 9887 91 - 1 CAUCCUCCGGUGUUCUCCCC------------CUUUGAGCAUUUUGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCCCAUCCUGUAAAUUGAAUGCAGA ........(((((((.....------------....))))))).........((((.(((((((......))))...))).))))(((((.......))))). ( -17.70) >DroSec_CAF1 9950 86 - 1 CCUCCCC-----UCCUCCCC------------UCAAGAGCAGUUGGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCUCAUCCUGAAAAUUGAAUGCAGG .......-----......((------------(((....(((((..((....((((((((((((......))))...)))))))).))..)))))..)).))) ( -20.40) >DroEre_CAF1 7772 89 - 1 CACCCCC-----UU---------CUCCGUCGCACAAGAGCAGUUGGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCUCAUCCUGAAAAUUGAAUGCAGG .......-----..---------.......(((......(((((..((....((((((((((((......))))...)))))))).))..)))))..)))... ( -20.20) >DroAna_CAF1 9000 78 - 1 GCUCCCC-----CACCC--------------------CGCCGUUGGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCUCAUCCUGAAAAUUGAAUGCAGG ((.....-----.....--------------------.(((...))).....((((((((((((......))))...)))))))).............))... ( -18.70) >DroPer_CAF1 9907 91 - 1 CAUCCUCCGGUAUUCUCCCC------------CUUUGAGCAUUUUGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCCCAUCCUGUAAAUUGAAUGCGGA .....(((((......))..------------......((((((((((....((((.(((((((......))))...))).)))).))))....))))))))) ( -19.30) >consensus CAUCCCC_____UUCUCCCC____________ACAAGAGCAGUUGGCAAAAUGAUGAGCUCAACAUAAAAGUUGCAUGGCUCAUCCUGAAAAUUGAAUGCAGG ......................................((.....)).....((((((((((((......))))...))))))))(((...........))). (-14.42 = -14.62 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:44 2006