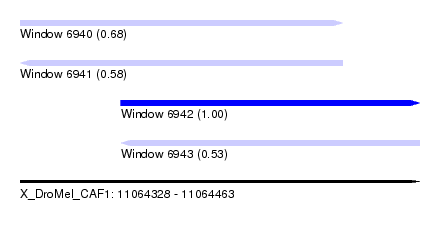
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,064,328 – 11,064,463 |
| Length | 135 |
| Max. P | 0.999257 |

| Location | 11,064,328 – 11,064,437 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11064328 109 + 22224390 AUCGGCAUACAA-AU-----GCGCCCACUGCGAUUCCAUUAGUCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAAC-----UUUUCCACUUCAACUUGGCCAGUUGGAUCACUUUGGCCAA ....((((.(.(-((-----(((((....((((((.....))))))..))).)))))..).))))...((((((.-----...((((((((.....))..)).)))).....)))))).. ( -31.10) >DroSec_CAF1 16516 109 + 1 AUCGGCAUACAA-AU-----GCGCCCACUGCGAUUCCAUUAGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAAC-----UUUUCCACUUCAACUUGGCCAGUUGGACCACUUUGGCCAA ...(((......-..-----.(((.....)))......(((((((....(((((.....))).))..))))))).-----..........(((..(((((....)).)))..)))))).. ( -29.30) >DroSim_CAF1 18181 109 + 1 AUCGGCAUACAA-AU-----GCGCCCACUGCGAUUCCAUUAGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAAC-----UUUUCCACUUCAACUUGGCCAGUUGGACCACUUUGGCCAA ...(((......-..-----.(((.....)))......(((((((....(((((.....))).))..))))))).-----..........(((..(((((....)).)))..)))))).. ( -29.30) >DroEre_CAF1 19808 109 + 1 AUCGGCAUACAA-AU-----GCGCCCAUUGCGAUUCCAUUAGCCGCUCGGCUGCAUUCUGCAUGCAUCGGCUAAC-----UUUUCCACUUCAACUUGGCCAGUUGGACCACAUUAGCCGU ....((((.(((-((-----(((((....(((...........)))..))).))))).)).))))..(((((((.-----........(((((((.....))))))).....))))))). ( -30.04) >DroYak_CAF1 17098 114 + 1 AUCGGCAUACAA-AUGCAAUGCGGCCAUUGCGAUUCCAUUAGCCGCUCGGCUGCAUUCUGCAUGCAUCGGCUAAC-----UUUUCCACUUCAACUUGGCCAGUUGCAGCACUUUAGCCAU ...(((......-(((((((((((((...(((...........)))..))))))))..)))))(((..((((((.-----..............))))))...))).........))).. ( -36.16) >DroPer_CAF1 46054 101 + 1 AUCCGCUGGCAAAAU-----AUGCU--CUUCCAAUCCAUUUACCGCUCUGGUGCACUCUGAAUUCC-AGGCACACACAUAUUUUGCAAC---------CCAUUUCGGCCACU--AUCCGA ........(((((((-----(((..--.....((.....))...((.((((..(.....)....))-)))).....))))))))))...---------.....((((.....--..)))) ( -18.50) >consensus AUCGGCAUACAA_AU_____GCGCCCACUGCGAUUCCAUUAGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAAC_____UUUUCCACUUCAACUUGGCCAGUUGGACCACUUUAGCCAA ...(((................................(((((((....(((((.....))).))..)))))))..............(((((((.....)))))))........))).. (-16.74 = -17.93 + 1.20)

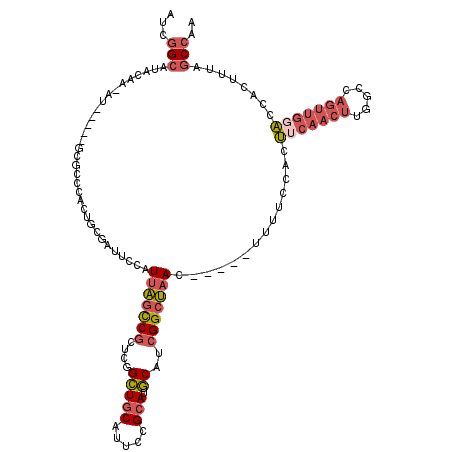
| Location | 11,064,328 – 11,064,437 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.60 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11064328 109 - 22224390 UUGGCCAAAGUGAUCCAACUGGCCAAGUUGAAGUGGAAAA-----GUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGACUAAUGGAAUCGCAGUGGGCGC-----AU-UUGUAUGCCGAU (((((((..((......)))))))))(((((..(....).-----.)))))((..(((((((.(((((.(((.(((((.........))))..).)))))-----))-)))))))))).. ( -36.10) >DroSec_CAF1 16516 109 - 1 UUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAA-----GUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGGCUAAUGGAAUCGCAGUGGGCGC-----AU-UUGUAUGCCGAU (((((..((((((((((.(((.(((........)))....-----((((((((.(((((.....)))))......)))))))).......)))))))).)-----))-))....))))). ( -38.30) >DroSim_CAF1 18181 109 - 1 UUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAA-----GUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGGCUAAUGGAAUCGCAGUGGGCGC-----AU-UUGUAUGCCGAU (((((..((((((((((.(((.(((........)))....-----((((((((.(((((.....)))))......)))))))).......)))))))).)-----))-))....))))). ( -38.30) >DroEre_CAF1 19808 109 - 1 ACGGCUAAUGUGGUCCAACUGGCCAAGUUGAAGUGGAAAA-----GUUAGCCGAUGCAUGCAGAAUGCAGCCGAGCGGCUAAUGGAAUCGCAAUGGGCGC-----AU-UUGUAUGCCGAU .((((((((....((((.((...........))))))...-----))))))))..(((((((((.(((.(((.((((..(.....)..)))..).)))))-----))-)))))))).... ( -39.90) >DroYak_CAF1 17098 114 - 1 AUGGCUAAAGUGCUGCAACUGGCCAAGUUGAAGUGGAAAA-----GUUAGCCGAUGCAUGCAGAAUGCAGCCGAGCGGCUAAUGGAAUCGCAAUGGCCGCAUUGCAU-UUGUAUGCCGAU .(((((((...(((.(((((.....))))).)))......-----.)))))))..(((((((((.(((((....(((((((.((......)).))))))).))))))-)))))))).... ( -43.20) >DroPer_CAF1 46054 101 - 1 UCGGAU--AGUGGCCGAAAUGG---------GUUGCAAAAUAUGUGUGUGCCU-GGAAUUCAGAGUGCACCAGAGCGGUAAAUGGAUUGGAAG--AGCAU-----AUUUUGCCAGCGGAU ((((..--.....)))).....---------(((((((((((((((((..(((-(.....))).)..))).....((((......))))....--.))))-----)))))).)))).... ( -28.40) >consensus UUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAA_____GUUAGCCGAUGCAUGCAGAAUGCAGCCGAGCGGCUAAUGGAAUCGCAGUGGGCGC_____AU_UUGUAUGCCGAU ..(.(((..(((((.(((((.....)))))...............((((((((.(((((.....)))))......))))))))...)))))..))).)...................... (-18.72 = -20.60 + 1.88)



| Location | 11,064,362 – 11,064,463 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11064362 101 + 22224390 AGUCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGGAUCACUUUGGCCAACCGGAAACCUACAAAAGUUGGCUGCA ((((....))))((((.....))))..((((((((((((....(((...((((((((..(.....).))))))))...)))......)))))))))))).. ( -31.20) >DroSec_CAF1 16550 101 + 1 AGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGGACCACUUUGGCCAACCGGAAACCUACAAAAGUUGGCUGCU ((((....))))((((.....))))..(((((((((((((((........)))((.(((((.((.....))))))).))........)))))))))))).. ( -34.20) >DroSim_CAF1 18215 101 + 1 AGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGGACCACUUUGGCCAACCGGAAACCUACAAAAGUUGGCUGCU ((((....))))((((.....))))..(((((((((((((((........)))((.(((((.((.....))))))).))........)))))))))))).. ( -34.20) >DroEre_CAF1 19842 101 + 1 AGCCGCUCGGCUGCAUUCUGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGGACCACAUUAGCCGUCCGGAAACCCACGAAAGUUGGCUGCC ..(((..(((((((((.....))))...((((((((...(((........)))..)))))).)).....)))))..)))...((.((....)).))..... ( -31.70) >DroYak_CAF1 17137 101 + 1 AGCCGCUCGGCUGCAUUCUGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGCAGCACUUUAGCCAUCCGGAAACCUACAAAAGUUGCUUGCU (((.(((.(((((.....(((.((((..((((((...............))))))...)))))))...)))))....(....).......))).))).... ( -24.96) >consensus AGCCGCUCGGCUGCAUUCCGCAUGCAUCGGCUAACUUUUCCACUUCAACUUGGCCAGUUGGACCACUUUGGCCAACCGGAAACCUACAAAAGUUGGCUGCU ((((....))))((((.....))))..((((((((((((....(((...((((((((..........))))))))...)))......)))))))))))).. (-28.56 = -28.92 + 0.36)


| Location | 11,064,362 – 11,064,463 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -28.00 |
| Energy contribution | -29.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11064362 101 - 22224390 UGCAGCCAACUUUUGUAGGUUUCCGGUUGGCCAAAGUGAUCCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGACU .((.((.(((((((.((..((((.(.(((((((..((......))))))))).).)))))).))))))).))((.(((((.....)))))..)).)).... ( -31.30) >DroSec_CAF1 16550 101 - 1 AGCAGCCAACUUUUGUAGGUUUCCGGUUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGGCU ...(((((((((((........((((((((((.....)).))))))))(((........)))))))))).((((.(((((.....)))))..)).)))))) ( -37.00) >DroSim_CAF1 18215 101 - 1 AGCAGCCAACUUUUGUAGGUUUCCGGUUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGGCU ...(((((((((((........((((((((((.....)).))))))))(((........)))))))))).((((.(((((.....)))))..)).)))))) ( -37.00) >DroEre_CAF1 19842 101 - 1 GGCAGCCAACUUUCGUGGGUUUCCGGACGGCUAAUGUGGUCCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCAGAAUGCAGCCGAGCGGCU ((.((((.((....)).)))).))...((((((((....((((.((...........))))))...)))))))).(((((.....)))))(((....))). ( -34.40) >DroYak_CAF1 17137 101 - 1 AGCAAGCAACUUUUGUAGGUUUCCGGAUGGCUAAAGUGCUGCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCAGAAUGCAGCCGAGCGGCU .....(((..(((((((.((...(((.(((((.....(((.(((((.....))))).))).....))))).)))..)).))))))))))((((....)))) ( -29.50) >consensus AGCAGCCAACUUUUGUAGGUUUCCGGUUGGCCAAAGUGGUCCAACUGGCCAAGUUGAAGUGGAAAAGUUAGCCGAUGCAUGCGGAAUGCAGCCGAGCGGCU ...(((((((((((.((..((((.(.(((((((..((......))))))))).).)))))).))))))).((((.(((((.....)))))..)).)))))) (-28.00 = -29.00 + 1.00)

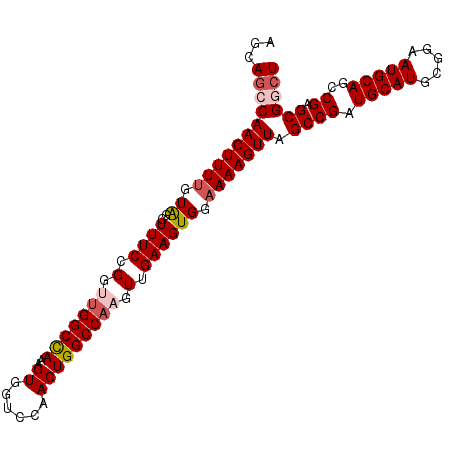
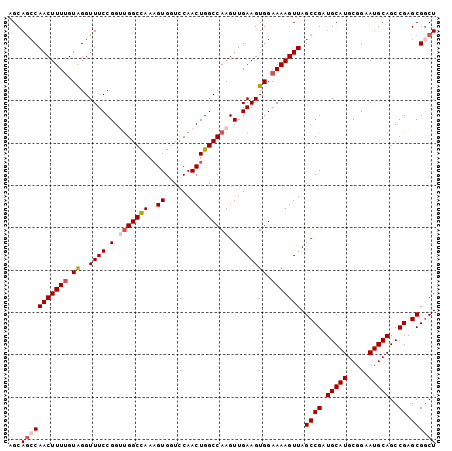
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:38 2006