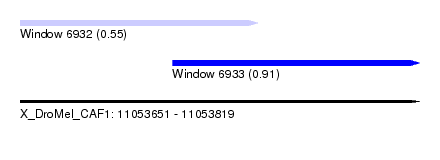
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,053,651 – 11,053,819 |
| Length | 168 |
| Max. P | 0.912200 |

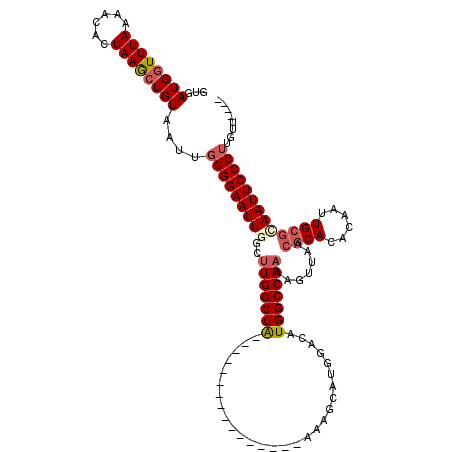
| Location | 11,053,651 – 11,053,751 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -23.23 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11053651 100 + 22224390 GUGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUGGCUUGGCCA----------------AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUU---- ...((((((((......))))))))(((.(((((((((..(((((((----------------............))))))).......((((......))))))))))))).)))---- ( -30.20) >DroSec_CAF1 5816 100 + 1 GUGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUGGCUUGGCCA----------------AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUUUU---- ...((((((((......))))))))....(((((((((..(((((((----------------............))))))).......((((......)))))))))))))....---- ( -29.70) >DroSim_CAF1 5096 100 + 1 GUGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUGGCUUGGCCA----------------AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUU---- ...((((((((......))))))))(((.(((((((((..(((((((----------------............))))))).......((((......))))))))))))).)))---- ( -30.20) >DroEre_CAF1 8040 100 + 1 GUGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUCGCUUGGCCA----------------AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGCU---- ...((((((((......))))))))....((((((((.(((((((((----------------............)))))))........(((......)))))))))))))....---- ( -28.00) >DroYak_CAF1 6412 100 + 1 GCGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUCGCUUGGCCA----------------AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUU---- ...((((((((......))))))))(((.((((((((.(((((((((----------------............)))))))........(((......))))))))))))).)))---- ( -28.50) >DroAna_CAF1 32358 117 + 1 GUAAUGCUUUAAUACACUAAACUGUAAUUGGGGAAUUGGCCUGGCCGACUGGUCGACUGGCCGACAGCAUG-ACUUGGCCAAAAGUCAACGCACACAGCUG--UAAUUCCCAGGACACAA ............((((......))))....((((((((((((((((....)))))...)))).(((((.((-((((......))))))..(....).))))--))))))))......... ( -34.10) >consensus GUGAUGGUUUAAAACACUAAGCUGUAAUUGGGGAAUUGGCUUGGCCA________________AAAGCAUGGACAUGGCCAAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUU____ ...((((((((......))))))))....(((((((((..(((((((............................))))))).......((((......)))))))))))))........ (-23.23 = -24.04 + 0.81)


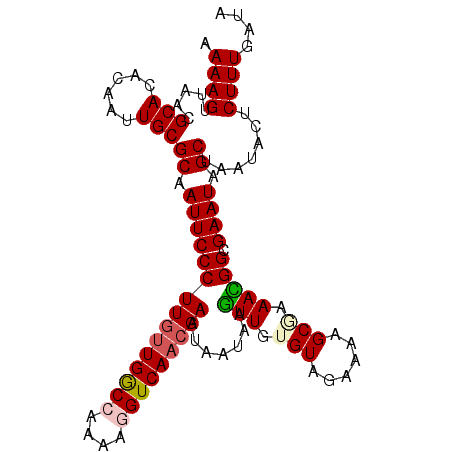
| Location | 11,053,715 – 11,053,819 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11053715 104 + 22224390 AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUUGGCCAAAAGGUCAACGAAUAAUAAGUUGUGUAGAAAAGCAAAACGGCGAAUAGCCUAUACUCUUU-UUA ...................(((((((((...(((((((((....)))))))))......)))))))))((((((......(((.....)))......))))-)). ( -25.90) >DroSec_CAF1 5880 105 + 1 AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUUUUGGCCAAAAGGUCAACGAAUAAUAAGUUGGGUAGAAAAGCGAAACGGCGAAUAGCUAAUACUCUUUGAUA ....(((((.(((......)))((.((((((...((((((....))))))..........(((..((......))..))))).)))).)).........))))). ( -19.20) >DroSim_CAF1 5160 105 + 1 AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUUGACCAAAAGGUCAACGAAUAAUAAGUUGGGUAGAAAAGCGAAACGGCGAAUAGCUAAUACUCUUUGAUA ....(((((.(((......)))((.(((((((((((((((....))))))))).......(((..((......))..))))).)))).)).........))))). ( -24.90) >DroEre_CAF1 8104 104 + 1 AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGCUGGC-AAAAGGUCAACGAAUAAUAAAUUGUGUAGAAAAGCGGAAUGGCGAAUAGCUGAUUCCCUUUGAAA ............((((((((..((((.....))))((((-.....))))..........))))))))...((((.((((((((.....))).))))))))).... ( -23.70) >DroYak_CAF1 6476 104 + 1 AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUUGGC-AAAAGGUCAACGAAUAAUAAGUUAUGUGGAAAAGCGAAAUGGCGAAUAGCUGACUCCCUUUGAAA ...(((((.(((.....(((.(((..((((.((((((((-.....))))))))((((....))))..))))..))).))).)))..))))).............. ( -21.50) >consensus AAAAGUUAACGCACACAAUUGCGCAAUUCCCUUGUUGGCCAAAAGGUCAACGAAUAAUAAGUUGUGUAGAAAAGCGAAACGGCGAAUAGCUAAUACUCUUUGAUA .((((.....(((......)))((.(((((((((((((((....))))))))).......(((.(((......))).))))).)))).)).......)))).... (-19.66 = -20.22 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:29 2006