
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,037,203 – 11,037,362 |
| Length | 159 |
| Max. P | 0.999994 |

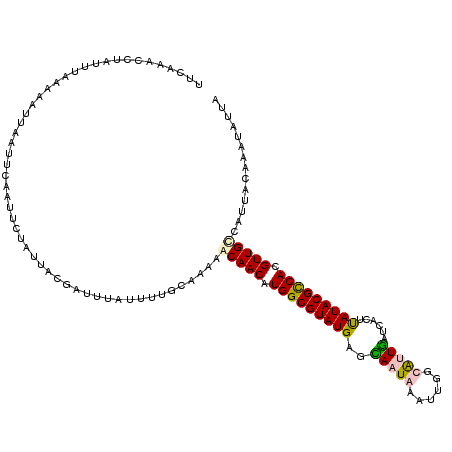
| Location | 11,037,203 – 11,037,323 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11037203 120 + 22224390 UUUAAAUCUAUUUAAAAAUUAAUUCAAUUCAUUUAUGAUUUAUUUUGCCAAAGCAACAUGGCGUAUGAGCAAUAAUUUGGUAAUGAUCACUUAUACGCCACGUUGUCAUUGCAAAUUUUA ..((((((............................)))))).(((((....(((((.(((((((((((..((.(((....))).))..))))))))))).)))))....)))))..... ( -27.89) >DroSec_CAF1 24454 113 + 1 UUUUAAUCUAUUUCAAAAUUAAUUCAAUUCUAUUAUGAUUUCUCUUGCCAAAGCAACAUGGCGUAUGCGCAAUAAUUUGGUAUUGAUCACCUAUACGUCACGUUGCCAUUAUA------- ....................................................(((((.(((((((((..(((((......)))))......))))))))).))))).......------- ( -19.00) >DroEre_CAF1 18699 120 + 1 UUCAAGCCUAUUUAAAAACUGAUUCAAUUCUAUAACGAUUUAUUUUGCAAAUACAACUUGGCGUAUGAGUAAUAAAUUCGCGCUGAUCACGUAUACGCCAUGUUGCUAUUACAAAUAUUA ...........................................((((..((((((((.((((((((((((.....))))(((.......))))))))))).)))).)))).))))..... ( -20.30) >DroYak_CAF1 19630 120 + 1 UGCAAACAUAUAUAAAUAUUAAUUCAAUUGUAUUACGAUUUACUUGGCAAAUACAACAUGGCGUAUGCGUAAUACAUUUUCAUUAAUCACUCAUACGCCAUGUUGCGGUCACAAAUAUUA ..............((((((.....(((((.....)))))....((((.....((((((((((((((.((.((............)).)).))))))))))))))..))))..)))))). ( -28.00) >consensus UUCAAACCUAUUUAAAAAUUAAUUCAAUUCUAUUACGAUUUAUUUUGCAAAAACAACAUGGCGUAUGAGCAAUAAAUUGGCAUUGAUCACUUAUACGCCACGUUGCCAUUACAAAUAUUA ....................................................(((((.(((((((((..(((((......)))))......))))))))).))))).............. (-15.74 = -16.18 + 0.44)

| Location | 11,037,203 – 11,037,323 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -23.91 |
| Energy contribution | -25.35 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 5.16 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11037203 120 - 22224390 UAAAAUUUGCAAUGACAACGUGGCGUAUAAGUGAUCAUUACCAAAUUAUUGCUCAUACGCCAUGUUGCUUUGGCAAAAUAAAUCAUAAAUGAAUUGAAUUAAUUUUUAAAUAGAUUUAAA .....(((((...(.(((((((((((((.((..((.(((....))).))..)).))))))))))))))....))))).((((((......((((((...)))))).......)))))).. ( -33.92) >DroSec_CAF1 24454 113 - 1 -------UAUAAUGGCAACGUGACGUAUAGGUGAUCAAUACCAAAUUAUUGCGCAUACGCCAUGUUGCUUUGGCAAGAGAAAUCAUAAUAGAAUUGAAUUAAUUUUGAAAUAGAUUAAAA -------......(((((((((.(((((..(((..(((((......))))))))))))).))))))))).......((....))..............(((((((......))))))).. ( -23.90) >DroEre_CAF1 18699 120 - 1 UAAUAUUUGUAAUAGCAACAUGGCGUAUACGUGAUCAGCGCGAAUUUAUUACUCAUACGCCAAGUUGUAUUUGCAAAAUAAAUCGUUAUAGAAUUGAAUCAGUUUUUAAAUAGGCUUGAA .....(((((((..(((((.((((((((..(((((.((......)).)))))..)))))))).)))))..))))))).(((..(.(((.(((((((...))))))))))...)..))).. ( -28.30) >DroYak_CAF1 19630 120 - 1 UAAUAUUUGUGACCGCAACAUGGCGUAUGAGUGAUUAAUGAAAAUGUAUUACGCAUACGCCAUGUUGUAUUUGCCAAGUAAAUCGUAAUACAAUUGAAUUAAUAUUUAUAUAUGUUUGCA ...((((((..(..(((((((((((((((.(((((..((....))..))))).)))))))))))))))..)..).)))))....((((.(((..(((((....)))))....))))))). ( -38.30) >consensus UAAUAUUUGUAAUGGCAACAUGGCGUAUAAGUGAUCAAUACCAAAUUAUUACGCAUACGCCAUGUUGCAUUGGCAAAAUAAAUCAUAAUAGAAUUGAAUUAAUUUUUAAAUAGAUUUAAA .....(((((((.(((((((((((((((..(((((.(((....))).)))))..))))))))))))))).))))))).......................(((((......))))).... (-23.91 = -25.35 + 1.44)

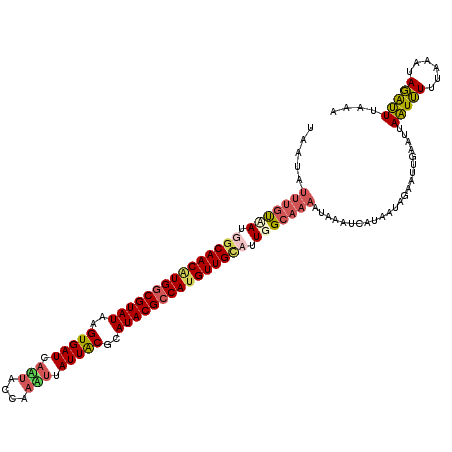
| Location | 11,037,243 – 11,037,362 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.16 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -17.67 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 5.84 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11037243 119 + 22224390 UAUUUUGCCAAAGCAACAUGGCGUAUGAGCAAUAAUUUGGUAAUGAUCACUUAUACGCCACGUUGUCAUUGCAAAUUUUAUGC-GACUCUAUUAUGGGGUAAUAUUUUUGCAUUUUUGCU ......((.((((((((.(((((((((((..((.(((....))).))..))))))))))).)))))...((((((...(((..-.(((((.....))))).)))..)))))).))).)). ( -33.80) >DroEre_CAF1 18739 108 + 1 UAUUUUGCAAAUACAACUUGGCGUAUGAGUAAUAAAUUCGCGCUGAUCACGUAUACGCCAUGUUGCUAUUACAAAUAUUAUAUUUGCUCCAUUAUUGGGUCAUAUAUU------------ ...((((..((((((((.((((((((((((.....))))(((.......))))))))))).)))).)))).))))....((((.((..(((....)))..)).)))).------------ ( -25.60) >DroYak_CAF1 19670 107 + 1 UACUUGGCAAAUACAACAUGGCGUAUGCGUAAUACAUUUUCAUUAAUCACUCAUACGCCAUGUUGCGGUCACAAAUAUUAUAC-GGCUCCAUUUUAGGGUUAAGCAGU------------ ..((((((.....((((((((((((((.((.((............)).)).)))))))))))))).((((.............-))))((......))))))))....------------ ( -28.72) >consensus UAUUUUGCAAAUACAACAUGGCGUAUGAGUAAUAAAUUCGCAAUGAUCACUUAUACGCCAUGUUGCCAUUACAAAUAUUAUAC_GGCUCCAUUAUAGGGUAAUAUAUU____________ .............((((.(((((((((.((.((............)).)).))))))))).))))....................((((.......)))).................... (-17.67 = -17.57 + -0.11)

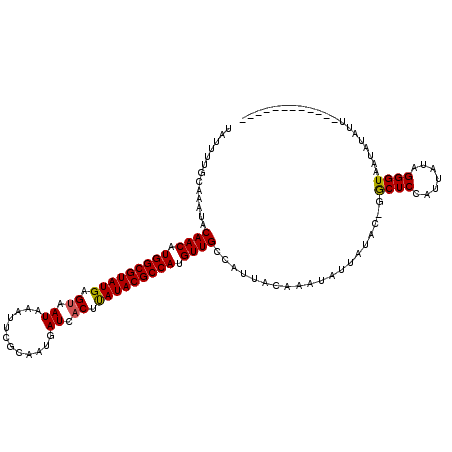

| Location | 11,037,243 – 11,037,362 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.16 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -26.58 |
| Energy contribution | -27.37 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.97 |
| Structure conservation index | 0.78 |
| SVM decision value | 5.86 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11037243 119 - 22224390 AGCAAAAAUGCAAAAAUAUUACCCCAUAAUAGAGUC-GCAUAAAAUUUGCAAUGACAACGUGGCGUAUAAGUGAUCAUUACCAAAUUAUUGCUCAUACGCCAUGUUGCUUUGGCAAAAUA .......((((.....(((((.....))))).....-))))....(((((...(.(((((((((((((.((..((.(((....))).))..)).))))))))))))))....)))))... ( -35.10) >DroEre_CAF1 18739 108 - 1 ------------AAUAUAUGACCCAAUAAUGGAGCAAAUAUAAUAUUUGUAAUAGCAACAUGGCGUAUACGUGAUCAGCGCGAAUUUAUUACUCAUACGCCAAGUUGUAUUUGCAAAAUA ------------.((((.((..(((....)))..)).))))....(((((((..(((((.((((((((..(((((.((......)).)))))..)))))))).)))))..)))))))... ( -30.40) >DroYak_CAF1 19670 107 - 1 ------------ACUGCUUAACCCUAAAAUGGAGCC-GUAUAAUAUUUGUGACCGCAACAUGGCGUAUGAGUGAUUAAUGAAAAUGUAUUACGCAUACGCCAUGUUGUAUUUGCCAAGUA ------------((.(((....((......))))).-))....((((((..(..(((((((((((((((.(((((..((....))..))))).)))))))))))))))..)..).))))) ( -37.20) >consensus ____________AAUAUAUAACCCAAUAAUGGAGCC_GUAUAAUAUUUGUAAUAGCAACAUGGCGUAUAAGUGAUCAAUGCAAAUUUAUUACUCAUACGCCAUGUUGUAUUUGCAAAAUA .............................................(((((((..((((((((((((((..(((((.(((....))).)))))..))))))))))))))..)))))))... (-26.58 = -27.37 + 0.79)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:19 2006