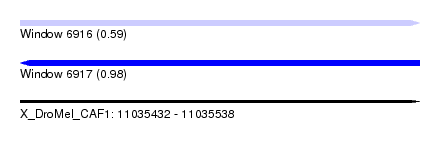
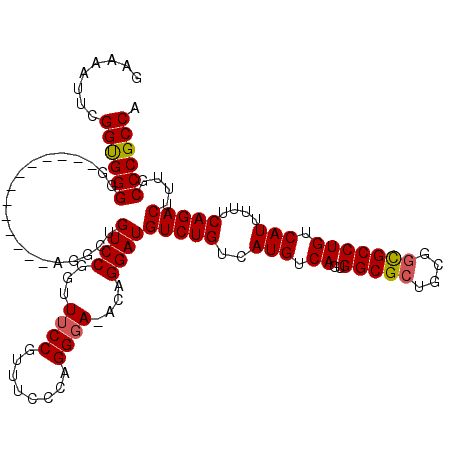
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,035,432 – 11,035,538 |
| Length | 106 |
| Max. P | 0.982095 |

| Location | 11,035,432 – 11,035,538 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11035432 106 + 22224390 UGGCGGGCAAAGUCUGAAAAAUGACAGGCACCGCAGCGCCACUGACAUGACAGACAUCCUGU-UCCCUGGGAAACGGAAACCGGACAGCCU-------------CCCCCACCGAAUUUUC .((.((((...(((((....(((.(((((........)))..)).)))..))))).(((.((-(.((..(....))).))).)))..))))-------------.))............. ( -28.90) >DroSec_CAF1 22757 107 + 1 UGGCGGGCAAAGUCUGAAGAAUGACAGGCGCCGCAGCGCCACUGACAUGACAGACAUCCUGUUUCCCUGGGAAACGGAAACCGGACAGCCU-------------CCCCCACCGAAUUUUC .((.((((...(((((....(((.(((((((....)))))..)).)))..))))).(((.((((((...(....))))))).)))..))))-------------.))............. ( -38.20) >DroSim_CAF1 14544 107 + 1 UGGCGGGCAAAGUCUGAAAAAUGACAGGCGCCGCAGCGCCACUGACAUGACAGACAUCCUGUUUCCCUGGGAAACGGAAACCGGACAGCCU-------------CCCCCACCGAAUUUUC .((.((((...(((((....(((.(((((((....)))))..)).)))..))))).(((.((((((...(....))))))).)))..))))-------------.))............. ( -38.20) >DroEre_CAF1 17032 106 + 1 UGGCGGGCAAAGUCUGAAAAAUGACAGGCGGCGCAGCGCCACUGACAUGACAGACAUCCUGU-UGCCUGGGACACGGAAGCCGGACAGCCU-------------CCCCCGCCGAAUUUUC .((((((....(((((....(((.(((..((((...)))).))).)))..))))).....((-((.((((....(....))))).))))..-------------..))))))........ ( -39.40) >DroYak_CAF1 17860 119 + 1 UGGCGGGCAAAGUCUGAAAAAUGACAGGCGCCGCAGCGCCACUGACAUGACAGACAUCCUGU-UCCCUGGGAAACGGAAACCGGACAGCCUCAGCCUCAGCCUCCCCCCACCGAAUUUUC .(((((((...(((((....(((.(((((((....)))))..)).)))..)))))...((((-((....(....)(....).)))))).....))))..))).................. ( -36.70) >consensus UGGCGGGCAAAGUCUGAAAAAUGACAGGCGCCGCAGCGCCACUGACAUGACAGACAUCCUGU_UCCCUGGGAAACGGAAACCGGACAGCCU_____________CCCCCACCGAAUUUUC .((.(((....(((((....(((.(((((((....)))))..)).)))..))))).(((.((.(((...(....)))).)).))).....................))).))........ (-29.38 = -29.82 + 0.44)

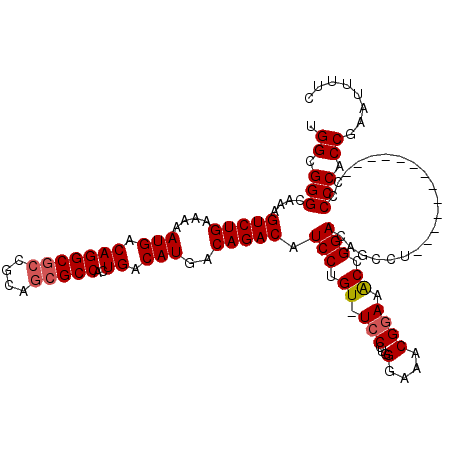
| Location | 11,035,432 – 11,035,538 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -47.26 |
| Consensus MFE | -37.74 |
| Energy contribution | -37.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11035432 106 - 22224390 GAAAAUUCGGUGGGGG-------------AGGCUGUCCGGUUUCCGUUUCCCAGGGA-ACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGGUGCCUGUCAUUUUUCAGACUUUGCCCGCCA ........((((((.(-------------((...((((.((((((........))))-)).))))(((((..(((.((..(((((....))))))).)))....)))))))).)))))). ( -43.90) >DroSec_CAF1 22757 107 - 1 GAAAAUUCGGUGGGGG-------------AGGCUGUCCGGUUUCCGUUUCCCAGGGAAACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGGCGCCUGUCAUUCUUCAGACUUUGCCCGCCA ........((((((((-------------(((((....)))))))((((((...)))))).....(((((..(((.((..(((((....))))))).)))....)))))....)))))). ( -45.40) >DroSim_CAF1 14544 107 - 1 GAAAAUUCGGUGGGGG-------------AGGCUGUCCGGUUUCCGUUUCCCAGGGAAACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGGCGCCUGUCAUUUUUCAGACUUUGCCCGCCA ........((((((((-------------(((((....)))))))((((((...)))))).....(((((..(((.((..(((((....))))))).)))....)))))....)))))). ( -45.40) >DroEre_CAF1 17032 106 - 1 GAAAAUUCGGCGGGGG-------------AGGCUGUCCGGCUUCCGUGUCCCAGGCA-ACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGCCGCCUGUCAUUUUUCAGACUUUGCCCGCCA ........((((((((-------------(((((....)))))))..((((..(...-.).))))(((((..(((.((((((((...))))).))).)))....)))))....)))))). ( -48.70) >DroYak_CAF1 17860 119 - 1 GAAAAUUCGGUGGGGGGAGGCUGAGGCUGAGGCUGUCCGGUUUCCGUUUCCCAGGGA-ACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGGCGCCUGUCAUUUUUCAGACUUUGCCCGCCA ........((((((.((((.(((((((..((((.((((.((((((........))))-)).)))))))))))(((.((..(((((....))))))).)))...)))).)))).)))))). ( -52.90) >consensus GAAAAUUCGGUGGGGG_____________AGGCUGUCCGGUUUCCGUUUCCCAGGGA_ACAGGAUGUCUGUCAUGUCAGUGGCGCUGCGGCGCCUGUCAUUUUUCAGACUUUGCCCGCCA ........((((((....................((((...((((........))))....))))(((((..(((.((..(((((....))))))).)))....)))))....)))))). (-37.74 = -37.82 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:13 2006