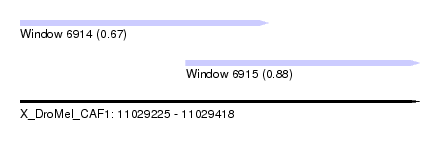
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,029,225 – 11,029,418 |
| Length | 193 |
| Max. P | 0.883215 |

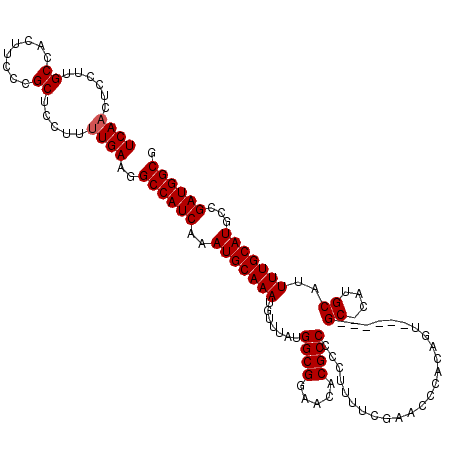
| Location | 11,029,225 – 11,029,345 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11029225 120 + 22224390 UCUGCUCGCCUCCAUCCGUGGCUACAAAUUUAUGGCGAGCCCUCGGCUCCGUUUGAUUGCGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCA ...(((((((........((....)).......)))))))....(((.....(((((...(((((......)))))...)))))......))).((((...........))))....... ( -25.76) >DroSec_CAF1 21640 111 + 1 UCUGCU---------CCGUGGCUACAAAUUUAUGGCGAGCCCUCGGCUCCGUUUGAUUGGGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACUUCCCGCCCCUUUUGAAGGCCAUCA ......---------..(((((..........(((((((((...)))))...(((((..((((((......))))))..)))))......))))((((...........))))))))).. ( -26.00) >DroSim_CAF1 13420 111 + 1 UCUGCU---------CCGUGGCUACAAAUUUAUGGCGAGCCCUCGGCUCCGUUUGAUUGCGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACUUCCCGCCCCUUUUGAAGGCCAUCA ......---------..(((((..........(((((((((...)))))...(((((...(((((......)))))...)))))......))))((((...........))))))))).. ( -25.50) >DroEre_CAF1 15875 117 + 1 U--GCUUGCC-CCAGCGGUGGCUACAAAUUUAUGGCGAGCCCUCGGCUCCGUUUGAUUGCGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACCUCCCGCUCCUUCUGAAGGCCAUCA .--(((....-..)))(((((((.........(((((((((...)))))...(((((...(((((......)))))...)))))......))))........((.....)).))))))). ( -29.00) >DroYak_CAF1 16690 104 + 1 C----------------GUGGCUACAAAUUUAUGGCGAGCCACCGGCUACGUUUGAUUGCGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCA .----------------(((((..........(((((((....((....)).(((((...(((((......)))))...)))))...)))))))((((...........))))))))).. ( -22.80) >consensus UCUGCU_________CCGUGGCUACAAAUUUAUGGCGAGCCCUCGGCUCCGUUUGAUUGCGAAUAUUAUCUUAUUUUAUGUCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCA .................(((((..........(((((((((...)))))...(((((...(((((......)))))...)))))......))))((((...........))))))))).. (-23.66 = -24.06 + 0.40)

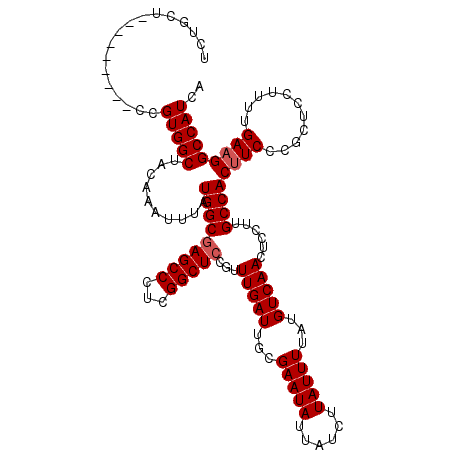

| Location | 11,029,305 – 11,029,418 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11029305 113 + 22224390 UCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCAAAUGCAAAUGUUUAUGGCGGAACACGCCCCCUUUGCGAACCCACAGU------GC-CAUGCAUUUUGCAUGCCGAUGGCG ..............((((...........))))((((((..((((((((((..(((((......(((.......))).((.....))------))-)))))).)))))))...)))))). ( -29.50) >DroSec_CAF1 21711 119 + 1 UCAACUCCUUGCCACUUCCCGCCCCUUUUGAAGGCCAUCAAAUGCAAAAGUUUAUGGCGGAACACGCCCCCUUUUCGAACCCACCAUGCAUUUGC-CAUGCAUUUUGCAUGCCGAUGGCG ..............((((...........))))((((((..((((((((......((((.....))))..................(((((....-.)))))))))))))...)))))). ( -31.20) >DroSim_CAF1 13491 119 + 1 UCAACUCCUUGCCACUUCCCGCCCCUUUUGAAGGCCAUCAAAUGCAAAUGUUUAUGGCGGAACACGCCCCCUUUUCGAAGCCACCAUGCAUUUGC-CAUGCAUUUUGCAUGCCGAUGGCG ..............((((...........))))((((((....((((((((..((((.((....((.........))...)).))))))))))))-(((((.....)))))..)))))). ( -33.50) >DroEre_CAF1 15952 111 + 1 UCAACUCCUUGCCACCUCCCGCUCCUUCUGAAGGCCAUCAAAUGCAAAUGUUUAUGGCGGAACACGCCCC--UUUUCGACCCACAGU------GC-CAUGCAUUUUGCAUGCCGAUGGCG (((.......((........))......)))..((((((..((((((((((..(((((((........))--......((.....))------))-)))))).)))))))...)))))). ( -26.82) >DroYak_CAF1 16754 114 + 1 UCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCAAAUGCAAAUGUUUAUGGCGGAACACGCCCCCUUUUCCGACCCACAGC------GCCCAUGCAUUUUGCAUGCCGAUGGCA ..............((((...........))))((((((..(((((((.(((...(((((((...........))))).))...)))------((....))..)))))))...)))))). ( -27.90) >consensus UCAACUCCUUGCCACUUCCCGCUCCUUUUGAAGGCCAUCAAAUGCAAAUGUUUAUGGCGGAACACGCCCCCUUUUCGAACCCACAGU______GC_CAUGCAUUUUGCAUGCCGAUGGCG ((((......((........)).....))))..((((((..(((((((.......((((.....)))).........................((....))..)))))))...)))))). (-25.30 = -25.50 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:11 2006