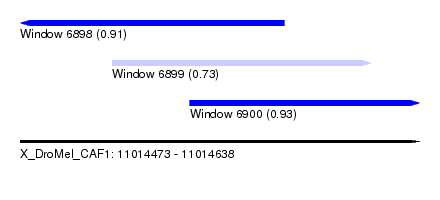
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,014,473 – 11,014,638 |
| Length | 165 |
| Max. P | 0.932662 |

| Location | 11,014,473 – 11,014,582 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -27.22 |
| Energy contribution | -27.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
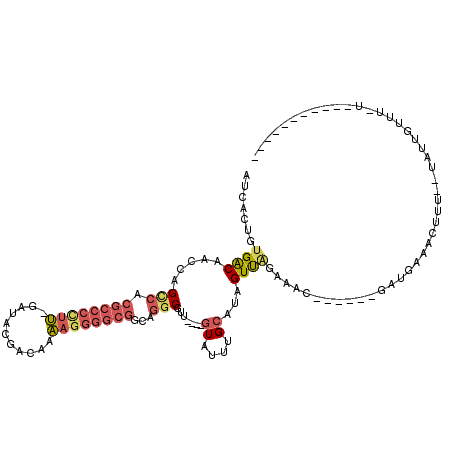
>X_DroMel_CAF1 11014473 109 - 22224390 UUUGUCGUAUCACAGGGCGUGGCUGAUUGUCACAGUGAUGAGAUGGCAAUUUCUCAACACGCUAACGAGAUGCACUUACAAAAAAAUGGGUAUGCCCAUU--GGGAAAUUA (((.(((((((.(..((((((((.....))))).(((.(((((........))))).))))))...).)))))...........((((((....))))))--)).)))... ( -34.60) >DroSec_CAF1 7337 108 - 1 UUUGUCGUAUCAAGGGGCGUGGCUGGUUGUCACAGUGAUGAGACGGCAAUUUCUCAACACGCUAACAAGAUGCACUCAAAA-AAAAUGGGUAUACUCAUU--CAGAAAUUA ((((..(((((....((((((((.....))))).(((.(((((........))))).)))))).....)))))...)))).-..((((((....))))))--......... ( -29.30) >DroSim_CAF1 2398 109 - 1 UUUGUCGUAUCAAGGGGCGUGGCUGGUUGUCACAGUGAUGAGACGGCAAUUUCUCAACACGCUAACAAGAUGCACUCAAAAAAAAAUGGGUAUACCCAUU--CGGAAAUUA ((((..(((((....((((((((.....))))).(((.(((((........))))).)))))).....)))))...))))....((((((....))))))--......... ( -32.30) >DroEre_CAF1 1039 98 - 1 UUUGUCGCAUCAAGGGGCGUGACCGACUGUCACAGUGGUGAGAUGGCAAUUUCUCAACACGCUAACAAGAUGCACUGAAAA--ACGGUGCUAUUUUUAAC----------- ......(((((....((((((((.....))))).(((.(((((........))))).)))))).....)))))((((....--.))))............----------- ( -28.80) >DroYak_CAF1 2345 109 - 1 UUUGUCGUAUCAAGGGGCGUGACUGGUUGUCACAGUGAUGAGACGGCAAUUUCUCAACACGCUAGCAAGAUGCACUGCAAG--AAAUGGGUAUUCUUAACGCCAAAAUCUA (((((.(((((....((((((((.....))))).(((.(((((........))))).)))))).....)))))...)))))--...(((((.......)).)))....... ( -28.70) >consensus UUUGUCGUAUCAAGGGGCGUGGCUGGUUGUCACAGUGAUGAGACGGCAAUUUCUCAACACGCUAACAAGAUGCACUCAAAA_AAAAUGGGUAUACUCAUU__CAGAAAUUA ......(((((....((((((((.....))))).(((.(((((........))))).)))))).....))))).............((((....))))............. (-27.22 = -27.06 + -0.16)


| Location | 11,014,511 – 11,014,618 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.98 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11014511 107 + 22224390 AUCUCGUUAGCGUGU-------UGAGAAAU-UGCCAUCUCAUCACUGUGACAAUCAGCCACGCCCUGU-GAUACGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUUAGAAAC .(((..((((((((.-------(((((...-.....))))).)))....(((((..(((.((((((.(-(......))...))))))...))))))----))....)).)))..)))... ( -32.70) >DroSec_CAF1 7374 107 + 1 AUCUUGUUAGCGUGU-------UGAGAAAU-UGCCGUCUCAUCACUGUGACAACCAGCCACGCCCCUU-GAUACGACAAAAGGGGCGGCGGGCGUU----GUAUUUGCUUAGUUAGAUAC ((((....((((((.-------(((((...-.....))))).)))....(((((..(((.((((((((-..........))))))))...))))))----))....))).....)))).. ( -39.00) >DroSim_CAF1 2436 107 + 1 AUCUUGUUAGCGUGU-------UGAGAAAU-UGCCGUCUCAUCACUGUGACAACCAGCCACGCCCCUU-GAUACGACAAAAGGGGCGGCGGGCGUU----GUAUUUGCUUAGUUAGAAAC .(((....((((((.-------(((((...-.....))))).)))....(((((..(((.((((((((-..........))))))))...))))))----))....))).....)))... ( -38.30) >DroEre_CAF1 1066 107 + 1 AUCUUGUUAGCGUGU-------UGAGAAAU-UGCCAUCUCACCACUGUGACAGUCGGUCACGCCCCUU-GAUGCGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUUAGCAAC ...(((((((((((.-------(((((...-.....))))).))).(((((.....)))))(((((((-..........))))))).(((((....----...)))))...)))))))). ( -37.60) >DroYak_CAF1 2383 107 + 1 AUCUUGCUAGCGUGU-------UGAGAAAU-UGCCGUCUCAUCACUGUGACAACCAGUCACGCCCCUU-GAUACGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUCGCAAAC ..((((((...(((.-------(((((...-.....))))).))).(((((.....)))))(((((((-..........)))))))))))))....----...(((((......))))). ( -38.20) >DroAna_CAF1 1088 110 + 1 AGAUUUCUAGCGUGUCUGGUGGUCACAAAUCCGCCAACUCAUCUCCCGGGCAAUCAGUCAUCCCCCUACCCUACCCCACUA----------UCUUUAUAAAUAUUAGCAUAGUUGGGAUC .((((((..(.(((..((((((........))))))...)))...)..)).))))..................(((.((((----------(((...........)).))))).)))... ( -21.80) >consensus AUCUUGUUAGCGUGU_______UGAGAAAU_UGCCAUCUCAUCACUGUGACAACCAGCCACGCCCCUU_GAUACGACAAAAGGGGCGGCAGGCGUU____GUAUUUGCAUAGUUAGAAAC .(((..(((((.(((.......(((((.........)))))........)))....(((.((((((((...........))))))))...))).............)).)))..)))... (-17.44 = -18.98 + 1.53)


| Location | 11,014,543 – 11,014,638 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.86 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -12.62 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11014543 95 + 22224390 AUCACUGUGACAAUCAGCCACGCCCUGU-GAUACGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUUAGAAAC------GAUGAAACUUG--UAUUGUUU-U----------- ...((((((.(((.(((((.((((((.(-(......))...))))))...)))..)----)...)))))))))..(((((------((((.......--))))))))-)----------- ( -27.90) >DroSec_CAF1 7406 96 + 1 AUCACUGUGACAACCAGCCACGCCCCUU-GAUACGACAAAAGGGGCGGCGGGCGUU----GUAUUUGCUUAGUUAGAUAC------GAUGAAACUUAU-UGUUUUUU-A----------- ........(((((...(((.((((((((-..........))))))))...)))(((----(((((((......)))))))------)))........)-))))....-.----------- ( -30.60) >DroSim_CAF1 2468 97 + 1 AUCACUGUGACAACCAGCCACGCCCCUU-GAUACGACAAAAGGGGCGGCGGGCGUU----GUAUUUGCUUAGUUAGAAAC------GAUGAAACUUUUGUUUUUUUU-U----------- ...((((..(((((..(((.((((((((-..........))))))))...))))))----)).......)))).((((((------((........))))))))...-.----------- ( -29.00) >DroEre_CAF1 1098 104 + 1 ACCACUGUGACAGUCGGUCACGCCCCUU-GAUGCGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUUAGCAAC------GU--CAGUUUU--UAUUGUUU-UUGGCCAAGCAG ....(((((.....)(((((((((((((-..........))))))))...((((((----((.............)))))------))--)......--........-.)))))..)))) ( -33.42) >DroYak_CAF1 2415 105 + 1 AUCACUGUGACAACCAGUCACGCCCCUU-GAUACGACAAAAGGGGCGGCAGGCGUU----GUAUUUGCAUAGUCGCAAAC------GA--AAGUUUU--UCUUGUUUUUUGGCCAAGUAC ...((((((((.....)))))(((((((-..........)))))))(((..(((.(----((....)))....)))((((------..--..)))).--............))).))).. ( -33.70) >DroAna_CAF1 1128 96 + 1 AUCUCCCGGGCAAUCAGUCAUCCCCCUACCCUACCCCACUA----------UCUUUAUAAAUAUUAGCAUAGUUGGGAUCUGAAAACAAAAGGCCCU--CAUUAUUU-U----------- .......((((..((((..(((((.(((..(((........----------.............)))..)))..)))))))))...(....))))).--........-.----------- ( -16.20) >consensus AUCACUGUGACAACCAGCCACGCCCCUU_GAUACGACAAAAGGGGCGGCAGGCGUU____GUAUUUGCAUAGUUAGAAAC______GAUGAAACUUU__UAUUGUUU_U___________ .......((((.....(((.((((((((...........))))))))...))).......((....))...))))............................................. (-12.62 = -13.52 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:56 2006