
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,006,493 – 11,006,587 |
| Length | 94 |
| Max. P | 0.908626 |

| Location | 11,006,493 – 11,006,587 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
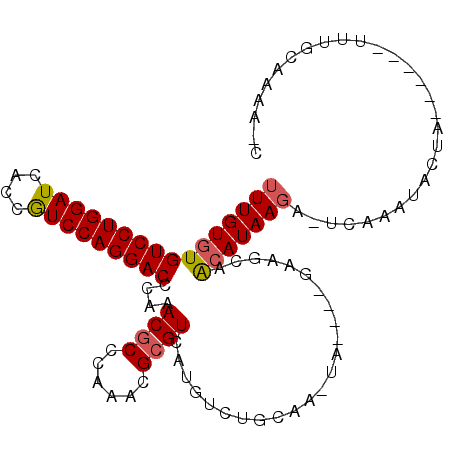
>X_DroMel_CAF1 11006493 94 + 22224390 G-UUUUGCAAC------UAGUAAUUGA-UCUUAUGUUGCUUC----UA-UUGCAGACAUGACGCGUUUAGGCGUUUGGGUCCUGGACGGUGAUCCAGGACACACAAA .-.(((((((.------.((((((...-......))))))..----..-)))))))((.(((((......))))))).((((((((......))))))))....... ( -29.60) >DroPse_CAF1 17109 102 + 1 GAUAGAAUAAAGAAUUCUACUAUUUCUUUAUUAUUCU-AUUC----UAUUUGCAGACAUGACGCGUUUAGGUGUUUGGGUCCUGGAUGGUGAUCCAGGACACACAAA ((((((((((((((.........))))...)))))))-)))(----((..(((.........)))..)))((((....(((((((((....)))))))))))))... ( -30.10) >DroSec_CAF1 7607 94 + 1 G-UUUUGCAAU------UAGUAAUUGA-UCUUAUGUUGCUUC----UA-UUGCAGACAUGACGCGCUUGGGCGUUUGGGUCCUGGACGGUGAUCCAGGACACACAAA .-.((((((((------.((((((...-......))))))..----.)-)))))))((.(((((......))))))).((((((((......))))))))....... ( -31.70) >DroSim_CAF1 4447 94 + 1 G-UUUUGCAAU------UAGUAAUUGA-UCUUAUGUUGCUUC----UA-UUGCAGACAUGACGCGCUUGGGCGUUUGGGUCCUGGACGGCGAUCCAGGACACACAAA .-.((((((((------.((((((...-......))))))..----.)-)))))))((.(((((......))))))).((((((((......))))))))....... ( -31.70) >DroEre_CAF1 6352 94 + 1 G-UAUUAUAAA------UAGUAUUUAA-UCUUAUGUUGAUUC----CA-UUGUAGACAUGACGCGUUUGGGCGUUUGGGUCCUGGAUGGCGAUCCAGGACACACAAA (-(((((....------))))))....-...((((((.((..----..-..)).))))))((((......))))(((.(((((((((....)))))))))...))). ( -24.80) >DroYak_CAF1 4463 98 + 1 A-AAUUACAAA------UAGUAUUUAA-UCUUAUGUUGAUUCAUUUCA-UUACAGACAUGACGCGUUUGGGCGUUUGGGUCCUGGACGGCGAUCCAGGACACACAAA .-.....(((.------..(((.....-...))).)))..((((.((.-.....)).))))(((......)))((((.((((((((......))))))))...)))) ( -22.20) >consensus G_UAUUACAAA______UAGUAAUUGA_UCUUAUGUUGAUUC____UA_UUGCAGACAUGACGCGUUUGGGCGUUUGGGUCCUGGACGGCGAUCCAGGACACACAAA ........................................................((.(((((......))))))).((((((((......))))))))....... (-20.29 = -20.15 + -0.14)



| Location | 11,006,493 – 11,006,587 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11006493 94 - 22224390 UUUGUGUGUCCUGGAUCACCGUCCAGGACCCAAACGCCUAAACGCGUCAUGUCUGCAA-UA----GAAGCAACAUAAGA-UCAAUUACUA------GUUGCAAAA-C .(((((.(((((((((....))))))))).)..((((......)))).......))))-..----...(((((.(((..-....)))...------)))))....-. ( -24.90) >DroPse_CAF1 17109 102 - 1 UUUGUGUGUCCUGGAUCACCAUCCAGGACCCAAACACCUAAACGCGUCAUGUCUGCAAAUA----GAAU-AGAAUAAUAAAGAAAUAGUAGAAUUCUUUAUUCUAUC ...(((((((((((((....)))))))))....))))(((...(((.......)))...))----).((-(((((((...((((.........))))))))))))). ( -24.00) >DroSec_CAF1 7607 94 - 1 UUUGUGUGUCCUGGAUCACCGUCCAGGACCCAAACGCCCAAGCGCGUCAUGUCUGCAA-UA----GAAGCAACAUAAGA-UCAAUUACUA------AUUGCAAAA-C ((((((.(((((((((....))))))))).)..((((......)))).((((.(((..-..----...)))))))....-..........------...))))).-. ( -24.50) >DroSim_CAF1 4447 94 - 1 UUUGUGUGUCCUGGAUCGCCGUCCAGGACCCAAACGCCCAAGCGCGUCAUGUCUGCAA-UA----GAAGCAACAUAAGA-UCAAUUACUA------AUUGCAAAA-C ((((((.(((((((((....))))))))).)..((((......)))).((((.(((..-..----...)))))))....-..........------...))))).-. ( -24.50) >DroEre_CAF1 6352 94 - 1 UUUGUGUGUCCUGGAUCGCCAUCCAGGACCCAAACGCCCAAACGCGUCAUGUCUACAA-UG----GAAUCAACAUAAGA-UUAAAUACUA------UUUAUAAUA-C ((((((((((((((((....)))))))))(((.((((......))))..((....)).-))----).....))))))).-..........------.........-. ( -23.80) >DroYak_CAF1 4463 98 - 1 UUUGUGUGUCCUGGAUCGCCGUCCAGGACCCAAACGCCCAAACGCGUCAUGUCUGUAA-UGAAAUGAAUCAACAUAAGA-UUAAAUACUA------UUUGUAAUU-U ((((((((((((((((....))))))))).....(((......)))((((.((.....-.)).))))....))))))).-.....(((..------...)))...-. ( -24.80) >consensus UUUGUGUGUCCUGGAUCACCGUCCAGGACCCAAACGCCCAAACGCGUCAUGUCUGCAA_UA____GAAGCAACAUAAGA_UCAAAUACUA______UUUGCAAAA_C ((((((((((((((((....)))))))))....((((......))))........................)))))))............................. (-20.14 = -20.28 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:50 2006