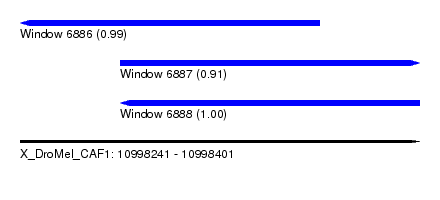
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,998,241 – 10,998,401 |
| Length | 160 |
| Max. P | 0.999552 |

| Location | 10,998,241 – 10,998,361 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -42.84 |
| Energy contribution | -42.44 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10998241 120 - 22224390 GGACGACAACAUGGAUCGCUGUGCGGACACCAUCUACAACUAUAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCGCGAGGAGCUGGUCCAGUGCACCCCAGAUGAU ((....((...(((((((((.(.((.(((((.(((.........))).))))).)..(((((..(((((((....)))).)))..)))))).).))).)))))).))....))....... ( -41.90) >DroSec_CAF1 10561 120 - 1 GGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCGCGAGGAGCUGGUCCAGUGCACCUGCGAUGAU ........((.(((((((((.(.((.(((((.(((.........))).))))).)..(((((..(((((((....)))).)))..)))))).).))).))))))))((....))...... ( -43.90) >DroSim_CAF1 12597 120 - 1 GGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCGCGAGUAGCUGGUCCAGUGCACCUGUGAUGAU ((..(.((...((((((((((..((.(((((.(((.........))).))))).)..(((((..(((((((....)))).)))..))))))..)))).)))))).))).))......... ( -45.30) >DroEre_CAF1 9862 120 - 1 UGAUGACAACAUGGACCGCUGUGCGGACACCAUCUACAAUUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCACGAGGAGCUGGUCCAGUGCACCUCCGAUGAU ...((.((...(((((((((.(.((.(((((.(((.........))).))))).)..(((((..(((((((....)))).)))..)))))).).))).)))))).))))........... ( -44.20) >DroYak_CAF1 10681 120 - 1 GGAUGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCACGAGGAGCUGGUCCAUUGCACCACCGAUGAU ((.((.((..((((((((((.(.((.(((((.(((.........))).))))).)..(((((..(((((((....)))).)))..)))))).).))).)))))))))))))......... ( -46.50) >consensus GGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGUGCAACGGCGCGAGGAGCUGGUCCAGUGCACCUCCGAUGAU ....(.((...((((((((((((...(((((.(((.........))).))))).)))(((((..(((((((....)))).)))..)))))....))).)))))).)))............ (-42.84 = -42.44 + -0.40)

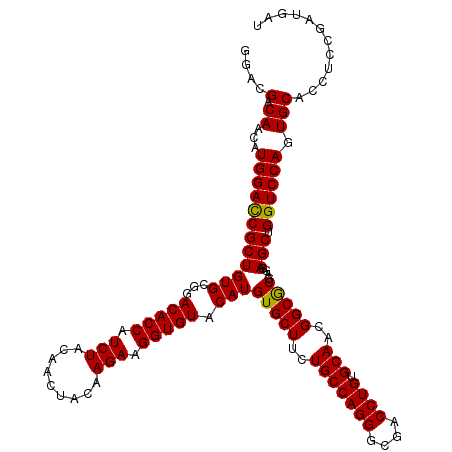
| Location | 10,998,281 – 10,998,401 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -47.30 |
| Consensus MFE | -47.28 |
| Energy contribution | -46.92 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10998281 120 + 22224390 ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUAUAGUUGUAGAUGGUGUCCGCACAGCGAUCCAUGUUGUCGUCCGGACCGCGCUGCACGCACAUCCGCUGGAUGGCCAGGUCGU ......((.((((((.(.(((.(((.((((.((......)).)))).)))(((((((.((.(((((....))))).)).)))).)))))).).....(((((...))))))))))).)). ( -43.20) >DroSec_CAF1 10601 120 + 1 ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUGUAGUUGUAGAUGGUGUCCGCACAGCGGUCCAUGUUGUCGUCCGGACCGCGCUGCACGCACAUCCGCUGGAUGGCCAGGUCGU ......((.((((((....((....(.(((((.(((.........))).))))).)(((..((((((((((....)))..)))))))..)))..)).(((((...))))))))))).)). ( -48.10) >DroSim_CAF1 12637 120 + 1 ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUGUAGUUGUAGAUGGUGUCCGCACAGCGGUCCAUGUUGUCGUCCGGACCGCGCUGCACGCACAUCCGCUGGAUGGCCAGGUCGU ......((.((((((....((....(.(((((.(((.........))).))))).)(((..((((((((((....)))..)))))))..)))..)).(((((...))))))))))).)). ( -48.10) >DroEre_CAF1 9902 120 + 1 ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUGUAAUUGUAGAUGGUGUCCGCACAGCGGUCCAUGUUGUCAUCAGGGCCGCGCUGCACGCACAUCCGCUGGGUGGCCAGGUCGU ...((((((((.(((.((.((....(.(((((.(((.........))).))))).)(((..(((((((.((.......)))))))))..)))..))...)).)))))))))))....... ( -48.60) >DroYak_CAF1 10721 120 + 1 ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUGUAGUUGUAGAUGGUGUCCGCACAGCGGUCCAUGUUGUCAUCCGGACCGCGCUGCACGCACAUCCGCUGGAUGGCCAGGUCGU ......((.((((((....((....(.(((((.(((.........))).))))).)(((..((((((((((....)))..)))))))..)))..)).(((((...))))))))))).)). ( -48.50) >consensus ACAGGUCGCCCUGGCAGAAGCACAUGUACACCUUCUUGUAGUUGUAGAUGGUGUCCGCACAGCGGUCCAUGUUGUCGUCCGGACCGCGCUGCACGCACAUCCGCUGGAUGGCCAGGUCGU ......((.((((((....((....(.(((((.(((.........))).))))).)(((..((((((((((....)))..)))))))..)))..)).(((((...))))))))))).)). (-47.28 = -46.92 + -0.36)

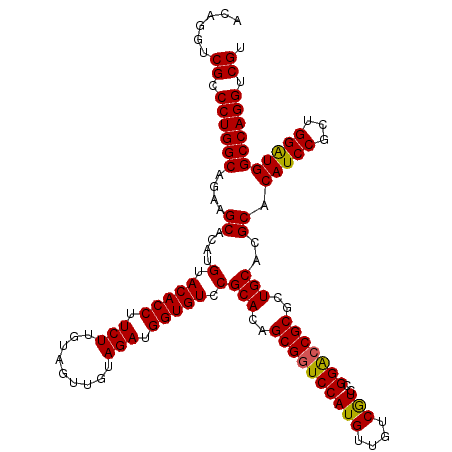
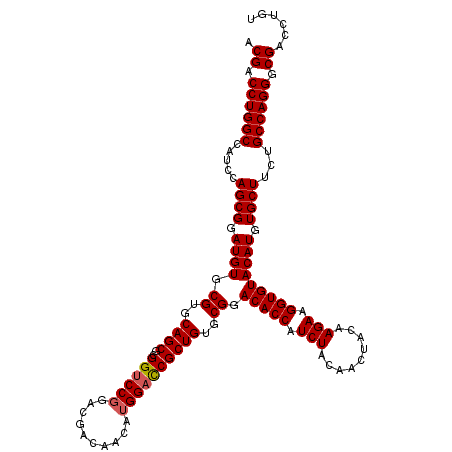
| Location | 10,998,281 – 10,998,401 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -49.74 |
| Consensus MFE | -48.66 |
| Energy contribution | -48.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10998281 120 - 22224390 ACGACCUGGCCAUCCAGCGGAUGUGCGUGCAGCGCGGUCCGGACGACAACAUGGAUCGCUGUGCGGACACCAUCUACAACUAUAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..(((((..(((((..........))))))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... ( -48.60) >DroSec_CAF1 10601 120 - 1 ACGACCUGGCCAUCCAGCGGAUGUGCGUGCAGCGCGGUCCGGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..((((..((((((..........))))))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... ( -50.80) >DroSim_CAF1 12637 120 - 1 ACGACCUGGCCAUCCAGCGGAUGUGCGUGCAGCGCGGUCCGGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..((((..((((((..........))))))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... ( -50.80) >DroEre_CAF1 9902 120 - 1 ACGACCUGGCCACCCAGCGGAUGUGCGUGCAGCGCGGCCCUGAUGACAACAUGGACCGCUGUGCGGACACCAUCUACAAUUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..(((((.(.((.((.......)).)).))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... ( -47.60) >DroYak_CAF1 10721 120 - 1 ACGACCUGGCCAUCCAGCGGAUGUGCGUGCAGCGCGGUCCGGAUGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..((((..((((((..(....)..))))))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... ( -50.90) >consensus ACGACCUGGCCAUCCAGCGGAUGUGCGUGCAGCGCGGUCCGGACGACAACAUGGACCGCUGUGCGGACACCAUCUACAACUACAAGAAGGUGUACAUGUGCUUCUGCCAGGGCGACCUGU .((.((((((.....((((.((((.((..((((..((((((..........))))))))))..)).(((((.(((.........))).))))))))).))))...)))))).))...... (-48.66 = -48.90 + 0.24)


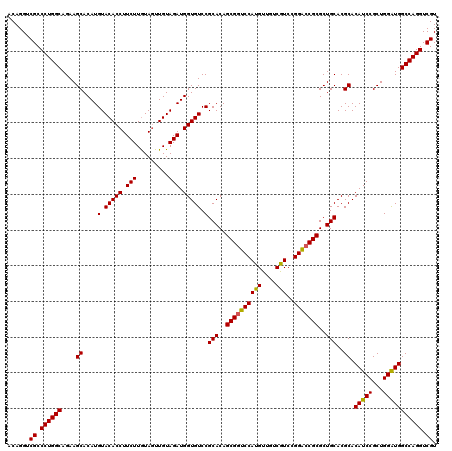
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:45 2006