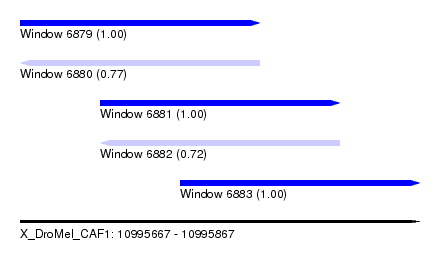
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,995,667 – 10,995,867 |
| Length | 200 |
| Max. P | 0.999281 |

| Location | 10,995,667 – 10,995,787 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10995667 120 + 22224390 UUCAAUAUUUUUCCAACGUGCAUCCUUAUCCAUCAAUCUCAACCUCAUUUGUGAUUGAAAUGUAUUCCCUUUUUUGCUCAGUGCUUGUGGUCAUUAGCGUUUUUAAUGGCCAUUAGCACC .................(.(((....(((...((((((.(((......))).))))))...)))..........))).).(((((.(((((((((((.....))))))))))).))))). ( -28.74) >DroSec_CAF1 7896 120 + 1 UUCAAUAUUUUCCCAAAGUGCGUCCUUAUCCAUCAAUACCAACCUCAAUUGUAAUUGAAAUGUAUUCCCUGUUUUGAUCAGUGCUAAUGGCCAUUAUCGUUUUUGAUGGCCAUUAGCACC .((((.((........(((((((.........(((((..(((......)))..))))).)))))))....)).))))...((((((((((((((((.......)))))))))))))))). ( -31.20) >DroSim_CAF1 9420 120 + 1 UUCAAUACUUUCCCAACGUGCGUCCUUAUCCAUCCAUACCAACCCCAAUUGUAAUUGAAAUGUAUUCUCUGUUUUGCUCAGUGCUAAUGGUCAUUAUCGUUUUUGAUGGCCAUUAGCACC ...(((((((((......((((...........................))))...)))).)))))..............((((((((((((((((.......)))))))))))))))). ( -27.03) >DroEre_CAF1 7084 112 + 1 UUUAAUAUUUUCCCUCCGUGCGUC-UUACCCAUC-------ACCUCAUAUUUAAUUAAUAUGCAUACCUUUUUUGGCUCAGUGCUUUUGGCCAUGAGUGGUUUUCAUUGCCAUUAGCACC ..................((.(((-.........-------....((((((.....))))))............))).))(((((..((((.(((((.....))))).))))..))))). ( -24.41) >DroYak_CAF1 7943 112 + 1 UUCCAUAUUU-ACCUCCGUGCGUCCUUACCCACC-------ACCUCAAUUUUAAUUAAUAUAUAUACCCUAUUUUGCUCUGUGCUUCUGGCCAUAAGCGGUUUUAAUGGCCAUUAGCACC ..........-......(((.((........)))-------)).....................................(((((..(((((((...........)))))))..))))). ( -19.80) >consensus UUCAAUAUUUUCCCAACGUGCGUCCUUAUCCAUC_AU__CAACCUCAAUUGUAAUUGAAAUGUAUUCCCUGUUUUGCUCAGUGCUUAUGGCCAUUAGCGUUUUUAAUGGCCAUUAGCACC ................................................................................(((((.(((((((((((.....))))))))))).))))). (-19.68 = -20.24 + 0.56)


| Location | 10,995,667 – 10,995,787 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -14.60 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10995667 120 - 22224390 GGUGCUAAUGGCCAUUAAAAACGCUAAUGACCACAAGCACUGAGCAAAAAAGGGAAUACAUUUCAAUCACAAAUGAGGUUGAGAUUGAUGGAUAAGGAUGCACGUUGGAAAAAUAUUGAA ((((((..(((.(((((.......))))).)))..))))))..(((.............(((((((((........))))))))).............)))................... ( -24.27) >DroSec_CAF1 7896 120 - 1 GGUGCUAAUGGCCAUCAAAAACGAUAAUGGCCAUUAGCACUGAUCAAAACAGGGAAUACAUUUCAAUUACAAUUGAGGUUGGUAUUGAUGGAUAAGGACGCACUUUGGGAAAAUAUUGAA (((((((((((((((...........)))))))))))))))..((((..((...(((((((((((((....))))))))..)))))..))..(((((.....)))))........)))). ( -34.20) >DroSim_CAF1 9420 120 - 1 GGUGCUAAUGGCCAUCAAAAACGAUAAUGACCAUUAGCACUGAGCAAAACAGAGAAUACAUUUCAAUUACAAUUGGGGUUGGUAUGGAUGGAUAAGGACGCACGUUGGGAAAGUAUUGAA (((((((((((.(((...........))).))))))))))).............(((((.((((.....((((..(.(((..(((......)))..))).)..)))).)))))))))... ( -28.40) >DroEre_CAF1 7084 112 - 1 GGUGCUAAUGGCAAUGAAAACCACUCAUGGCCAAAAGCACUGAGCCAAAAAAGGUAUGCAUAUUAAUUAAAUAUGAGGU-------GAUGGGUAA-GACGCACGGAGGGAAAAUAUUAAA ((((((..((((.((((.......)))).))))..))))))..(((......)))...((((((.....))))))..((-------(.((.....-..)))))................. ( -24.90) >DroYak_CAF1 7943 112 - 1 GGUGCUAAUGGCCAUUAAAACCGCUUAUGGCCAGAAGCACAGAGCAAAAUAGGGUAUAUAUAUUAAUUAAAAUUGAGGU-------GGUGGGUAAGGACGCACGGAGGU-AAAUAUGGAA .(((((..(((((((...........)))))))..))))).............((((.(((.(((((....)))))...-------.(((.((....)).)))....))-).)))).... ( -25.40) >consensus GGUGCUAAUGGCCAUCAAAAACGCUAAUGGCCAUAAGCACUGAGCAAAAAAGGGAAUACAUUUCAAUUACAAUUGAGGUUG__AU_GAUGGAUAAGGACGCACGUUGGGAAAAUAUUGAA ((((((..(((((((...........)))))))..))))))..................((((((((....))))))))......................................... (-14.60 = -16.00 + 1.40)


| Location | 10,995,707 – 10,995,827 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10995707 120 + 22224390 AACCUCAUUUGUGAUUGAAAUGUAUUCCCUUUUUUGCUCAGUGCUUGUGGUCAUUAGCGUUUUUAAUGGCCAUUAGCACCUGUGCAGCUUCCACCUAACAAUCCCAGCCACACCUAAUGU .....((((((((..((...(((..........((((.(((((((.(((((((((((.....))))))))))).)))).))).))))..........)))....))..))))...)))). ( -31.25) >DroSec_CAF1 7936 120 + 1 AACCUCAAUUGUAAUUGAAAUGUAUUCCCUGUUUUGAUCAGUGCUAAUGGCCAUUAUCGUUUUUGAUGGCCAUUAGCACCUGUGCUGAUUCCACCUAACAAUCCCAGCCACGCCUAAUGU ....(((((....)))))......................((((((((((((((((.......))))))))))))))))..((((((.................))).)))......... ( -32.73) >DroSim_CAF1 9460 120 + 1 AACCCCAAUUGUAAUUGAAAUGUAUUCUCUGUUUUGCUCAGUGCUAAUGGUCAUUAUCGUUUUUGAUGGCCAUUAGCACCUGUGCUGAUUCCACCUAACAAUCCCAGCCACGCCUAAAAU ........(((..((((..................((.((((((((((((((((((.......))))))))))))))).))).))((....)).....))))..)))............. ( -30.30) >DroEre_CAF1 7117 116 + 1 -ACCUCAUAUUUAAUUAAUAUGCAUACCUUUUUUGGCUCAGUGCUUUUGGCCAUGAGUGGUUUUCAUUGCCAUUAGCACCUGUG---CUUCCACCUCACAAUCACCGAAACACUUAAUGU -....((((((.....))))))........((((((....(((((..((((.(((((.....))))).))))..))))).((((---.........))))....)))))).......... ( -26.80) >DroYak_CAF1 7976 116 + 1 -ACCUCAAUUUUAAUUAAUAUAUAUACCCUAUUUUGCUCUGUGCUUCUGGCCAUAAGCGGUUUUAAUGGCCAUUAGCACCUGUG---AUUCCACCUCACAAUUAGCUAGACACUUAAUGU -..................................(((..(((((..(((((((...........)))))))..))))).((((---(.......)))))...))).............. ( -23.70) >consensus AACCUCAAUUGUAAUUGAAAUGUAUUCCCUGUUUUGCUCAGUGCUUAUGGCCAUUAGCGUUUUUAAUGGCCAUUAGCACCUGUGC_GAUUCCACCUAACAAUCCCAGCCACACCUAAUGU ........................................(((((.(((((((((((.....))))))))))).)))))......................................... (-19.68 = -20.24 + 0.56)



| Location | 10,995,707 – 10,995,827 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10995707 120 - 22224390 ACAUUAGGUGUGGCUGGGAUUGUUAGGUGGAAGCUGCACAGGUGCUAAUGGCCAUUAAAAACGCUAAUGACCACAAGCACUGAGCAAAAAAGGGAAUACAUUUCAAUCACAAAUGAGGUU ((.(((..((((..((..((.((...........(((...((((((..(((.(((((.......))))).)))..))))))..)))...........))))..))..))))..))).)). ( -31.05) >DroSec_CAF1 7936 120 - 1 ACAUUAGGCGUGGCUGGGAUUGUUAGGUGGAAUCAGCACAGGUGCUAAUGGCCAUCAAAAACGAUAAUGGCCAUUAGCACUGAUCAAAACAGGGAAUACAUUUCAAUUACAAUUGAGGUU .........(((.(((.((((.....(((.......))).(((((((((((((((...........)))))))))))))))))))....)))....)))((((((((....)))))))). ( -35.70) >DroSim_CAF1 9460 120 - 1 AUUUUAGGCGUGGCUGGGAUUGUUAGGUGGAAUCAGCACAGGUGCUAAUGGCCAUCAAAAACGAUAAUGACCAUUAGCACUGAGCAAAACAGAGAAUACAUUUCAAUUACAAUUGGGGUU .........(((.(((.((((.(.....).)))).((...(((((((((((.(((...........))).)))))))))))..))....)))....)))((..((((....))))..)). ( -30.50) >DroEre_CAF1 7117 116 - 1 ACAUUAAGUGUUUCGGUGAUUGUGAGGUGGAAG---CACAGGUGCUAAUGGCAAUGAAAACCACUCAUGGCCAAAAGCACUGAGCCAAAAAAGGUAUGCAUAUUAAUUAAAUAUGAGGU- .......(((((((.(...........).))))---))).((((((..((((.((((.......)))).))))..))))))..(((......)))...((((((.....))))))....- ( -30.00) >DroYak_CAF1 7976 116 - 1 ACAUUAAGUGUCUAGCUAAUUGUGAGGUGGAAU---CACAGGUGCUAAUGGCCAUUAAAACCGCUUAUGGCCAGAAGCACAGAGCAAAAUAGGGUAUAUAUAUUAAUUAAAAUUGAGGU- ..............(((..((((((.......)---)))))(((((..(((((((...........)))))))..)))))..)))..................................- ( -28.10) >consensus ACAUUAGGUGUGGCUGGGAUUGUUAGGUGGAAUC_GCACAGGUGCUAAUGGCCAUCAAAAACGCUAAUGGCCAUAAGCACUGAGCAAAAAAGGGAAUACAUUUCAAUUACAAUUGAGGUU .................(((((....(((...........((((((..(((((((...........)))))))..))))))...............)))....)))))............ (-17.87 = -17.79 + -0.08)



| Location | 10,995,747 – 10,995,867 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -22.52 |
| Energy contribution | -24.68 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10995747 120 + 22224390 GUGCUUGUGGUCAUUAGCGUUUUUAAUGGCCAUUAGCACCUGUGCAGCUUCCACCUAACAAUCCCAGCCACACCUAAUGUGACACUCACGUUCUUGUCGUCAUUAGGAUCCAGUGCAGAU (((((.(((((((((((.....))))))))))).)))))(((..(.(((................)))....((((((((((((..........))))).))))))).....)..))).. ( -40.59) >DroSec_CAF1 7976 120 + 1 GUGCUAAUGGCCAUUAUCGUUUUUGAUGGCCAUUAGCACCUGUGCUGAUUCCACCUAACAAUCCCAGCCACGCCUAAUGUGACACUCACGCUCUUGCCAUCAUUAGGAUCCAAUACAGAU ((((((((((((((((.......))))))))))))))))((((((((.................))))....(((((((((.....)))((....))....)))))).......)))).. ( -42.43) >DroSim_CAF1 9500 120 + 1 GUGCUAAUGGUCAUUAUCGUUUUUGAUGGCCAUUAGCACCUGUGCUGAUUCCACCUAACAAUCCCAGCCACGCCUAAAAUGACACUCACGCUCUUGCCAUCANNNNNNNNNNNNNNNNNN ((((((((((((((((.......))))))))))))))))..((((((.................))).)))..................((....))....................... ( -29.13) >DroEre_CAF1 7156 117 + 1 GUGCUUUUGGCCAUGAGUGGUUUUCAUUGCCAUUAGCACCUGUG---CUUCCACCUCACAAUCACCGAAACACUUAAUGUGACACUCACGCUCUUUCCACCAUUAGGAUCCCGUGCAGAU (((((..((((.(((((.....))))).))))..)))))(((..---(..............................(((.....)))......(((.......)))....)..))).. ( -30.70) >DroYak_CAF1 8015 117 + 1 GUGCUUCUGGCCAUAAGCGGUUUUAAUGGCCAUUAGCACCUGUG---AUUCCACCUCACAAUUAGCUAGACACUUAAUGUGACACUCACGCUCUUGCCACCAUUAGGAUCCCGUGCAGAU (((((..(((((((...........)))))))..)))))(((((---..(((...(((((.((((........))))))))).......((....))........))).....))))).. ( -30.20) >consensus GUGCUUAUGGCCAUUAGCGUUUUUAAUGGCCAUUAGCACCUGUGC_GAUUCCACCUAACAAUCCCAGCCACACCUAAUGUGACACUCACGCUCUUGCCAUCAUUAGGAUCCAGUGCAGAU (((((.(((((((((((.....))))))))))).)))))(((((.....(((..........................(((.....)))((....))........))).....))))).. (-22.52 = -24.68 + 2.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:40 2006