
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,992,869 – 10,993,038 |
| Length | 169 |
| Max. P | 0.713876 |

| Location | 10,992,869 – 10,992,986 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10992869 117 + 22224390 GUAUUGGUGAGAACCUGUGGACGCUGCGAAGCGCGCCAA-AAAAUAAAAAUGCCGACAUACAAAUAGUGCAGCGUUUUC--UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGC .(((.(((....))).)))(((((((((..(..(((((.-......(((((((.(.(((.......)))).))))))).--(((((......))))).)))))..)..)))))).))).. ( -37.90) >DroSec_CAF1 4971 116 + 1 GUAUUGGUGAGAACCUGUGGACGCUGCGAAGCGCGGCAAAAAAAUAAAAAUGCCGACAUACAAAUAGUGCAGCGUUUUC--UUAGCUCAUUUGCUGGCUGGCAAACAG--CAGCAGUCGC ......(((((.....(..((((((((.....((((((............))))).)..((.....))))))))))..)--....)))))((((((.(((.....)))--)))))).... ( -36.90) >DroEre_CAF1 4650 116 + 1 GUAUUGGUGAGAACCUGUGGACGCUGCAAAGCGCGCCAA-AA---AAAAAUGCCGACAUACAAAUAAUGCAUCGUUUUUUGUUUGCUCAUUUGCUAGUUGGCGACGAGCGCAGCAGUCGC .(((.(((....))).)))((((((((....((((((((-..---..(((((..(.((.(((((.((((...)))).))))).))).))))).....)))))).))...))))).))).. ( -35.00) >DroYak_CAF1 5460 113 + 1 GCAUCGGUGAGAACCUGUGGACGCUGCAAAGCGCGCCAA-AA---AAAAAUGCCGACAUACAAAUAAUGCUGUGUUUUC---UUGCUCAUUUGCUGGCUGGCGACGAGCGCAGCAGUCGC .(((.(((....))).)))((((((((....(((((((.-..---......(((((((((((.....)).))))))...---..((......)).)))))))).))...))))).))).. ( -35.50) >consensus GUAUUGGUGAGAACCUGUGGACGCUGCAAAGCGCGCCAA_AA___AAAAAUGCCGACAUACAAAUAAUGCAGCGUUUUC__UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGC .(((.(((....))).)))((((((((......(((((........(((((((.(.(((.......)))).)))))))....((((......))))..)))))......))))).))).. (-26.69 = -27.38 + 0.69)

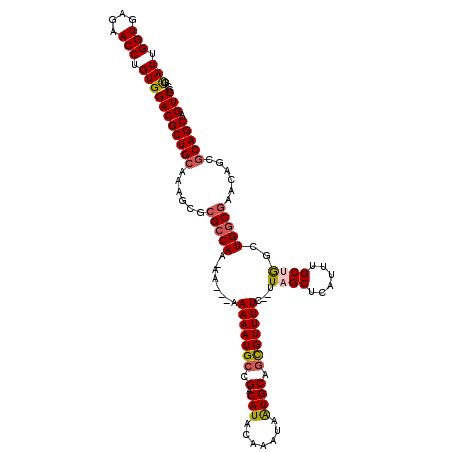
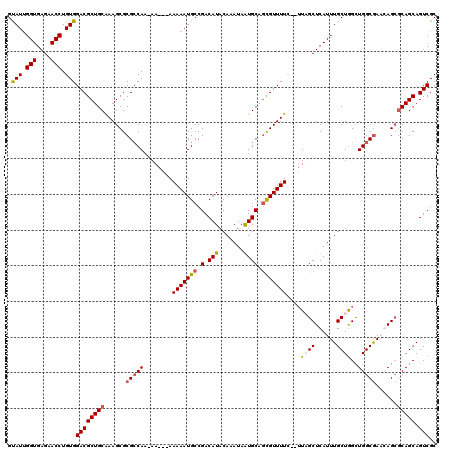
| Location | 10,992,908 – 10,993,026 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.29 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10992908 118 + 22224390 AAAAUAAAAAUGCCGACAUACAAAUAGUGCAGCGUUUUC--UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUUUAUAUAUUCUAA ...(((.((((((.(((........((((.(((......--...)))))))(((((((((.....)))).))))))))((((((........)))))).....)))))).)))....... ( -33.80) >DroSec_CAF1 5011 116 + 1 AAAAUAAAAAUGCCGACAUACAAAUAGUGCAGCGUUUUC--UUAGCUCAUUUGCUGGCUGGCAAACAG--CAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUUUAUAUAUUCUAA ...(((.((((((.(((........((((.(((......--...)))))))(((((.(((.....)))--))))))))((((((........)))))).....)))))).)))....... ( -30.50) >DroEre_CAF1 4689 116 + 1 AA---AAAAAUGCCGACAUACAAAUAAUGCAUCGUUUUUUGUUUGCUCAUUUGCUAGUUGGCGACGAGCGCAGCAGUCGCCAUUCACGUGAAAAUCGCCAAUCGCAUU-CUAAAUUCUAA ..---...(((((.((((.(((((.((((...)))).))))).)).)).((..(....(((((((..((...)).))))))).....)..))...........)))))-........... ( -24.90) >DroYak_CAF1 5499 112 + 1 AA---AAAAAUGCCGACAUACAAAUAAUGCUGUGUUUUC---UUGCUCAUUUGCUGGCUGGCGACGAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUU--UAAAUUCUAA ..---..((((((.(((..........((((((((((..---(((((..((....))..))))).)))))))))))))((((((........)))))).....)))))--)......... ( -28.80) >consensus AA___AAAAAUGCCGACAUACAAAUAAUGCAGCGUUUUC__UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUU_AUAAAUUCUAA .......((((((.(((...(((((...((..............))..)))))...((((((.....)).)))).)))((((((........)))))).....))))))........... (-20.48 = -21.29 + 0.81)

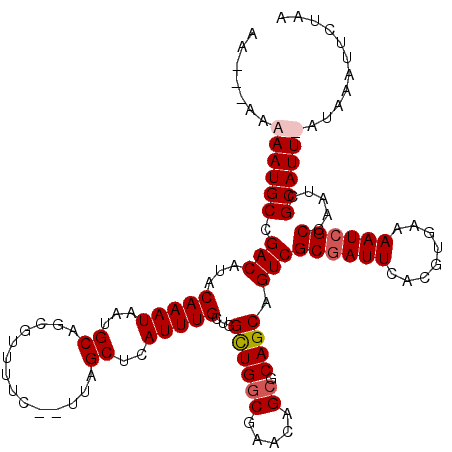
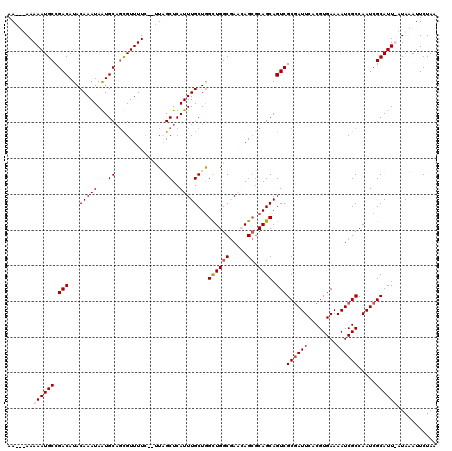
| Location | 10,992,908 – 10,993,026 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.69 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10992908 118 - 22224390 UUAGAAUAUAUAAAUGCGAUUGGCGAUUUUCACGUGAAUCGCGACUGCUGCGCUGUUCGCCAGCCAGCAAAUGAGCUAA--GAAAACGCUGCACUAUUUGUAUGUCGGCAUUUUUAUUUU ((((..(((.....(((((((.(((.......))).)))))))..(((((.((((.....))))))))).)))..))))--(((((.((((.((.........)))))).)))))..... ( -33.90) >DroSec_CAF1 5011 116 - 1 UUAGAAUAUAUAAAUGCGAUUGGCGAUUUUCACGUGAAUCGCGACUGCUG--CUGUUUGCCAGCCAGCAAAUGAGCUAA--GAAAACGCUGCACUAUUUGUAUGUCGGCAUUUUUAUUUU ((((..(((.....(((((((.(((.......))).)))))))..(((((--(((.....))).))))).)))..))))--(((((.((((.((.........)))))).)))))..... ( -30.60) >DroEre_CAF1 4689 116 - 1 UUAGAAUUUAG-AAUGCGAUUGGCGAUUUUCACGUGAAUGGCGACUGCUGCGCUCGUCGCCAACUAGCAAAUGAGCAAACAAAAAACGAUGCAUUAUUUGUAUGUCGGCAUUUUU---UU .........((-(((((..((((((((.(((....))).((((.(....))))).))))))))...(((((((((((............))).))))))))......))))))).---.. ( -29.00) >DroYak_CAF1 5499 112 - 1 UUAGAAUUUA--AAUGCGAUUGGCGAUUUUCACGUGAAUCGCGACUGCUGCGCUCGUCGCCAGCCAGCAAAUGAGCAA---GAAAACACAGCAUUAUUUGUAUGUCGGCAUUUUU---UU .........(--(((((((((.(((.......))).)))).((((.((((.((.....))))))..((((((((((..---.........)).))))))))..))))))))))..---.. ( -31.30) >consensus UUAGAAUAUAU_AAUGCGAUUGGCGAUUUUCACGUGAAUCGCGACUGCUGCGCUCGUCGCCAGCCAGCAAAUGAGCAAA__GAAAACGCUGCACUAUUUGUAUGUCGGCAUUUUU___UU ............(((((((((.(((.......))).)))).((((.((((.((.....))))))..((((((((((..............)).))))))))..)))))))))........ (-23.44 = -24.69 + 1.25)


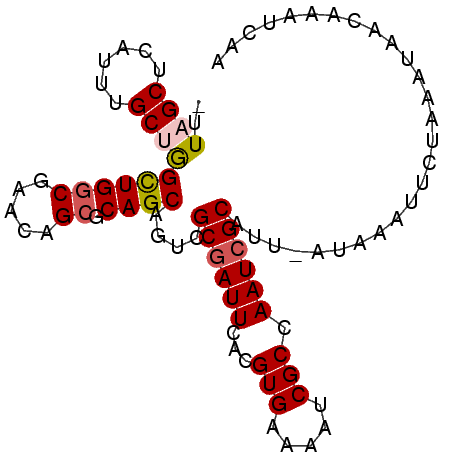
| Location | 10,992,947 – 10,993,038 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -15.77 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10992947 91 + 22224390 -UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUUUAUAUAUUCUAAAUAACAAAUCGA -((((.....((((((((((.....)))).))))))..((((((...(((.....))).))))))...........))))............ ( -24.20) >DroSec_CAF1 5050 89 + 1 -UUAGCUCAUUUGCUGGCUGGCAAACAG--CAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUUUAUAUAUUCUAAAUAACAAAUCAA -((((.....((((((.(((.....)))--))))))..((((((...(((.....))).))))))...........))))............ ( -20.90) >DroEre_CAF1 4726 91 + 1 GUUUGCUCAUUUGCUAGUUGGCGACGAGCGCAGCAGUCGCCAUUCACGUGAAAAUCGCCAAUCGCAUU-CUAAAUUCUAAAUAACAAAUCAA (..(((..(((.((....(((((((..((...)).)))))))(((....)))....)).))).)))..-)...................... ( -18.10) >DroYak_CAF1 5535 88 + 1 --UUGCUCAUUUGCUGGCUGGCGACGAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUU--UAAAUUCUAAAUAACAAAUCAA --........((((((((((....).))).))))))..((((((...(((.....))).))))))...--...................... ( -21.40) >consensus _UUAGCUCAUUUGCUGGCUGGCGAACAGCGCAGCAGUCGCGAUUCACGUGAAAAUCGCCAAUCGCAUU_AUAAAUUCUAAAUAACAAAUCAA ..((((......))))((((((.....)).))))....((((((...(((.....))).))))))........................... (-15.77 = -16.40 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:33 2006