
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,985,898 – 10,985,994 |
| Length | 96 |
| Max. P | 0.517495 |

| Location | 10,985,898 – 10,985,994 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 54.73 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -9.39 |
| Energy contribution | -9.62 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.30 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10985898 96 + 22224390 UUGGGUGAGAGGUGGCAGCCAGGUCGCGGCCAUGGCUGUUUGUCGCAGCUGGAAGGGAUUGGGCGAAAU-------------GGGAU---------AUAGUGA-GGGCAUGCGAUCAAA .(((.((........)).)))((((((((((.(.(((((..(((.(((((.((.....)).)))....)-------------).)))---------))))).)-.))).)))))))... ( -32.00) >DroPse_CAF1 8929 115 + 1 CCGGACGAAAGUCGGCAUCCAGGAAGCGGAAAAGGUUGCGGCUCACAGCUGUAG---AUGGAGCA-AAUCGAACGUUAGCUGUGUGUAAAACAUGAAUGAUGAAGGCCAGACAGCCAGC ...(((....)))(((.(((.(....))))....(((..((((((((((((..(---.(.((...-..)).).)..)))))))).......(((.....)))..)))).))).)))... ( -33.60) >DroEre_CAF1 10760 90 + 1 UUGGGGAAGAGAUGGCAGCCAGGUCGCGGCCAGGGCUGUUUGUCGCAGCUGGAAGGGAUU--GGG-----------------GGUCU---------AUAGUGU-UGGCUUUUGGCCAAA ((((..((((((..((((((.(((....)))..))))).)..))((.((((....((((.--...-----------------.))))---------.))))..-..))))))..)))). ( -27.90) >DroYak_CAF1 10411 92 + 1 UUGGGGGAGAGAUGGCAGCCAGGUCGCGGCCAGGGCUGUUUGUCGCAGCUGGAAGGGAUUAAGGG-----------------GGUCU---------AUAGUGA-UGGCUUUUGGCCAAA ((((..((((((..((((((.(((....)))..))))).)..))((.((((....(((((.....-----------------)))))---------.))))..-..))))))..)))). ( -28.80) >DroAna_CAF1 13540 85 + 1 CCGGCGGACAAGCGACAGCCGUUCCGUGGAAACGGAAGUGGGUCACAGCUGAAA----UUAU--------------------GCUUU---------GUUUUAA-GACCCAUUACUCCAU .(.(((((...((....))...))))).)....(((((((((((.....(((((----(...--------------------.....---------)))))).-))))))))..))).. ( -25.50) >DroPer_CAF1 8909 115 + 1 CCGGACGAAAGUCGGCAUCCAGGAAGCGGAAAAGGUUGCGGCUCACAGCUGUAG---AUGGAGAA-UAUCCAACGUUAGCUGUGUGUAAAACAUGAAUGAUGAAGGCCAGACAGCCAAC ...(((....)))(((.(((.(....))))....(((..((((((((((((..(---.((((...-..)))).)..)))))))).......(((.....)))..)))).))).)))... ( -38.00) >consensus CCGGGGGAGAGACGGCAGCCAGGUCGCGGAAAAGGCUGUGGGUCACAGCUGGAA___AUUGAGCG_________________GGUCU_________AUAGUGA_GGCCAUUCAGCCAAA .............(((.(((.......)))...((((((.....))))))...............................................................)))... ( -9.39 = -9.62 + 0.22)

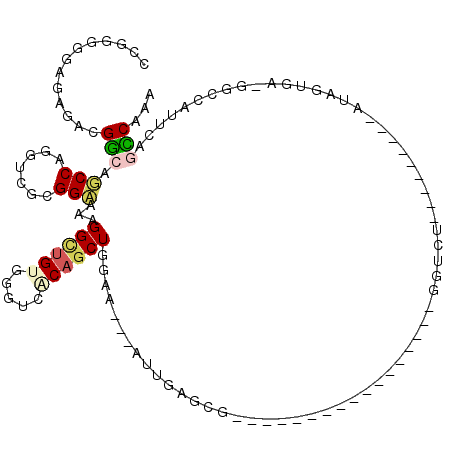
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:29 2006