
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,982,869 – 10,982,985 |
| Length | 116 |
| Max. P | 0.808290 |

| Location | 10,982,869 – 10,982,985 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.33 |
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -5.34 |
| Energy contribution | -5.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10982869 116 + 22224390 CUU----GUAGUUUUAUGGUUGUUAAUUAUCUUUUUUUCAAAAACAUUUAGCCAUUUACCAAUUUGGACAAAGAGCUUUCCACAUGUAUACAAAUUGCUUGUACAGUGGAAACACCCACA ..(----(((((((((((((((..........................)))))))...((.....))....))))))((((((.((((((.........)))))))))))))))...... ( -19.77) >DroSec_CAF1 6734 111 + 1 AUU----GU-GCUUAAUGGCUGUAAAUUAUCUCUUUUUCAAAAACAUUUAGCCAUUUAUCAAU--GGACAUAGAGCUGUCC--AUAUAUAAAAAUUGCUUGUACAGUGGAUACAUCCACA ..(----((-((..((((((((..........................)))))))).....((--(((((......)))))--))...............)))))(((((....))))). ( -27.37) >DroSim_CAF1 6776 115 + 1 AUU----GU-GCUUAAUGGCUUUAAAUUAUCUCUUUUUCAAAAACAUUUAGCCAUUUAUCAAUUUGGACAUAGAGCUGUCUAAAUAUAUAAAAAUUCCUUGUACAGUGGAUACAUCCACA ..(----((-((..(((((((............................))))))).....(((((((((......)))))))))...............)))))(((((....))))). ( -25.49) >DroEre_CAF1 7426 112 + 1 AUUUCCUUGAGAAACACGCAUUUAAGUUAUU-AUUUUUCGUAGGCAGUUAGCCAUUUAUCAAUUUGGACAAAGUGUUU---GC---UGUGUU-GUUGUUUGUUAAGUGGAAACACCCGCA .(((((((((.(((((((((..(((....))-)......(((((((.((.((((..........))).).)).)))))---))---))))).-...)))).))))).))))......... ( -21.70) >DroYak_CAF1 7066 118 + 1 AUUUCAUUCGGUUUUACACUUUUAAAUUUUG-GUUUUUUGGAAUCAUUUAACCAUUUAUCCAUUUGGACAAAGUUCUAUAUACAUAUGUGCAAAUUCUUUGU-CAGUGGAAACACCCUCA .........(((.................((-(((...((....))...)))))....(((((...(((((((.......(((....)))......))))))-).)))))...))).... ( -20.62) >consensus AUU____GU_GCUUAAUGGCUUUAAAUUAUCUCUUUUUCAAAAACAUUUAGCCAUUUAUCAAUUUGGACAAAGAGCUGUCCACAUAUAUAAAAAUUGCUUGUACAGUGGAAACACCCACA ...................................................(((..........)))......................................((((......)))). ( -5.34 = -5.62 + 0.28)

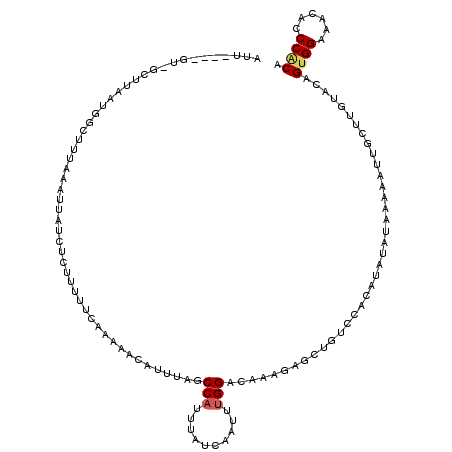
| Location | 10,982,869 – 10,982,985 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.33 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -7.48 |
| Energy contribution | -8.56 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10982869 116 - 22224390 UGUGGGUGUUUCCACUGUACAAGCAAUUUGUAUACAUGUGGAAAGCUCUUUGUCCAAAUUGGUAAAUGGCUAAAUGUUUUUGAAAAAAAGAUAAUUAACAACCAUAAAACUAC----AAG (((((.(((((((((((((((((...))))))))...))))))((((.((((.((.....)))))).))))...(((((((.....)))))))....))).))))).......----... ( -27.10) >DroSec_CAF1 6734 111 - 1 UGUGGAUGUAUCCACUGUACAAGCAAUUUUUAUAUAU--GGACAGCUCUAUGUCC--AUUGAUAAAUGGCUAAAUGUUUUUGAAAAAGAGAUAAUUUACAGCCAUUAAGC-AC----AAU .(((((....))))).......((......(((..((--(((((......)))))--))..)))(((((((((((.(((((....)))))...))))..)))))))..))-..----... ( -28.90) >DroSim_CAF1 6776 115 - 1 UGUGGAUGUAUCCACUGUACAAGGAAUUUUUAUAUAUUUAGACAGCUCUAUGUCCAAAUUGAUAAAUGGCUAAAUGUUUUUGAAAAAGAGAUAAUUUAAAGCCAUUAAGC-AC----AAU .(((((....)))))(((.(..........(((..((((.((((......)))).))))..)))(((((((((((.(((((....)))))...))))..)))))))..).-))----).. ( -22.50) >DroEre_CAF1 7426 112 - 1 UGCGGGUGUUUCCACUUAACAAACAAC-AACACA---GC---AAACACUUUGUCCAAAUUGAUAAAUGGCUAACUGCCUACGAAAAAU-AAUAACUUAAAUGCGUGUUUCUCAAGGAAAU ...(..((((.......))))..)...-(((((.---((---(.....((((((......)))))).(((.....)))..........-...........))))))))............ ( -17.10) >DroYak_CAF1 7066 118 - 1 UGAGGGUGUUUCCACUG-ACAAAGAAUUUGCACAUAUGUAUAUAGAACUUUGUCCAAAUGGAUAAAUGGUUAAAUGAUUCCAAAAAAC-CAAAAUUUAAAAGUGUAAAACCGAAUGAAAU ...((.((((......)-))).....(((((((...............(((((((....)))))))(((((...((....))...)))-))..........))))))).))......... ( -21.40) >consensus UGUGGGUGUUUCCACUGUACAAGCAAUUUGUAUAUAUGUAGAAAGCUCUUUGUCCAAAUUGAUAAAUGGCUAAAUGUUUUUGAAAAAGAGAUAAUUUAAAACCAUAAAAC_AC____AAU .(((((....)))))............................((((.((((((......)))))).))))................................................. ( -7.48 = -8.56 + 1.08)

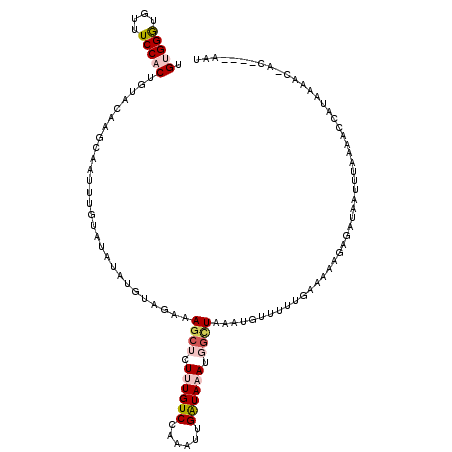
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:19 2006