
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,979,139 – 10,979,267 |
| Length | 128 |
| Max. P | 0.995814 |

| Location | 10,979,139 – 10,979,237 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.84 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10979139 98 - 22224390 GAUAACGUAGUCUCGUGUUGACAAAUCCAAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAAUACUCAGUAUUGGCAGAAAACAAUGCAACUUA ((((((((((((....(((((...((((....))))...)))))....))------------)....)))))))))........((((((........))))))...... ( -29.10) >DroSec_CAF1 2990 89 - 1 GAUAACGCACGCUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAAUCCUCA---------GAAAACAAUGCGACUUA (((((((((((.(((.(((((...((((....))))...)))))...)))------------...)))))))))))........---------................. ( -26.50) >DroSim_CAF1 3048 89 - 1 GAUAACGCACGCUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAAUCCUCA---------GAAAACAAUGCGACUUA (((((((((((.(((.(((((...((((....))))...)))))...)))------------...)))))))))))........---------................. ( -26.50) >DroEre_CAF1 3627 105 - 1 GAUAACGCA--CUCGUGUUGACAAAUCCUAACGAGUGAAUCAGCUGCUGAUUGG---CGGGGCAUUAUGCGUUAUCAAUCCUCAUUAAUAGUAGAAAACAAUGCGAUUUA (((((((((--(((((...((....))...)))))).......(((((....))---)))........))))))))..............(((........)))...... ( -26.10) >DroYak_CAF1 3279 98 - 1 GAUAACGCA--CUCGUGUUGACAAAUU-UAGCCAUC---------GAUGAUUGGUGGUGGUGCAUCAUGCGUUAUCAAUACUCAGUAAUAGUAGUAAACAAUGAGAUUUA (((((((((--...((((..((.....-..((((((---------((...)))))))).))))))..)))))))))....((((.................))))..... ( -23.93) >consensus GAUAACGCA__CUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA____________CAUCGUGCGUUAUCAAUCCUCA_UA_U_G_AGAAAACAAUGCGACUUA (((((((((.......(((((...((((....))))...)))))...(((..............)))))))))))).................................. (-12.96 = -13.84 + 0.88)


| Location | 10,979,171 – 10,979,267 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
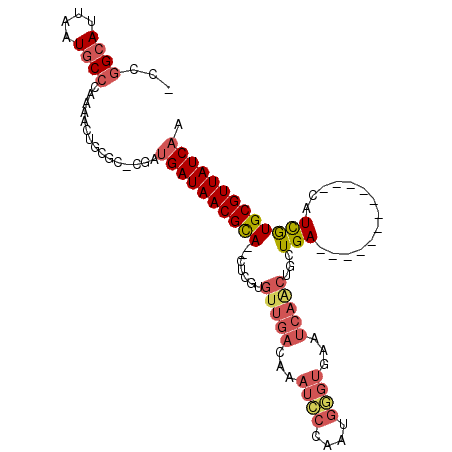
>X_DroMel_CAF1 10979171 96 - 22224390 CCCGGCAUUAAUGCACAAAAUCUCGC-CGAUGAUAACGUAGUCUCGUGUUGACAAAUCCAAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAA ..((((..................))-)).(((((((((((((....(((((...((((....))))...)))))....))------------)....)))))))))). ( -28.67) >DroSec_CAF1 3013 96 - 1 ACCGGCAUUAAUGCCCAAAACUGCGC-CGAUGAUAACGCACGCUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAA ..((((..................))-)).((((((((((((.(((.(((((...((((....))))...)))))...)))------------...)))))))))))). ( -32.37) >DroSim_CAF1 3071 96 - 1 ACCGGCAUUAGUGCCCAAAACUGCGC-CGAUGAUAACGCACGCUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA------------CAUCGUGCGUUAUCAA ..((((..((((.......)))).))-)).((((((((((((.(((.(((((...((((....))))...)))))...)))------------...)))))))))))). ( -35.80) >DroEre_CAF1 3659 103 - 1 -CCGGCAUUAAUACCCAAGACUAUGCACUAAGAUAACGCA--CUCGUGUUGACAAAUCCUAACGAGUGAAUCAGCUGCUGAUUGG---CGGGGCAUUAUGCGUUAUCAA -...((((..............)))).....(((((((((--(((((...((....))...)))))).......(((((....))---)))........)))))))).. ( -27.64) >DroYak_CAF1 3311 95 - 1 -CCGGCAUUAAUGCCGAAGACUUCGC-CGAUGAUAACGCA--CUCGUGUUGACAAAUU-UAGCCAUC---------GAUGAUUGGUGGUGGUGCAUCAUGCGUUAUCAA -.(((((....)))))..........-...((((((((((--...((((..((.....-..((((((---------((...)))))))).))))))..)))))))))). ( -30.00) >consensus _CCGGCAUUAAUGCCCAAAACUGCGC_CGAUGAUAACGCA__CUCGUGUUGACAAAUCCCAAUGGGUGAAUCAACUGCUGA____________CAUCGUGCGUUAUCAA ...((((....))))...............((((((((((.......(((((...((((....))))...)))))...(((..............))))))))))))). (-15.48 = -16.96 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:17 2006