
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,953,561 – 10,953,682 |
| Length | 121 |
| Max. P | 0.966245 |

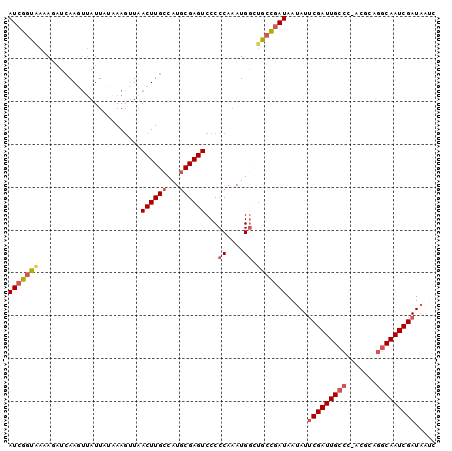
| Location | 10,953,561 – 10,953,660 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.28 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10953561 99 + 22224390 AUCGGCGAAAGCUCAAUUUAUUAUAAAUAUUACUUG-AAUGCGAGUCCCCCAUAUGGCUGCCGAUAAUAUUCGAUUGCCCGACGCAAGCAAUCGAUAAGC (((((((...((((.(((((..............))-)))..))))...((....)).))))))).....((((((((.........))))))))..... ( -23.84) >DroSec_CAF1 4199 100 + 1 AUCGGUAAAAUAUCAAGUUAUUCCAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCCUACGCAGGCAAUCGAUAUUC (((((((.......((((((.........))))))(((((.............))))))))))))(((((.((((((((.......))))))))))))). ( -30.52) >DroSim_CAF1 1798 100 + 1 AUCGGUAAAAUAUGAAGUUAUUCCAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCUUACGCAGGCAAUCGAUAUUC (((((((.......((((((.........))))))(((((.............))))))))))))(((((.(((((((((.....)))))))))))))). ( -32.12) >DroEre_CAF1 6756 99 + 1 AUCGGUAAAAGAUCAAGUGAUUAUAAAGUUAACUUGCUAUGCGAGUCCCCCAAAUGAAUGGUGAUAACAUGCGAUUGCCC-AGAAAGACAAUCGAUAACC ((((((....((((....)))).....(((.((((((...)))))).............((..((........))..)).-.....))).)))))).... ( -16.40) >DroYak_CAF1 3520 98 + 1 AUCGGUAAAAGAUCAAGCUAUUAUUAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGG-GCCUAUAAUAUUCGAUUGCCC-AGAAAGGCAAUCGAUAAUA .(..((((.((......)).))))..)((((((((((...)))))).((((....)))-)....))))..(((((((((.-.....)))))))))..... ( -26.40) >consensus AUCGGUAAAAGAUCAAGUUAUUAUAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCC_ACGCAGGCAAUCGAUAAUC (((((((........................((((((...))))))...((....)).))))))).....(((((((((.......)))))))))..... (-19.12 = -20.28 + 1.16)


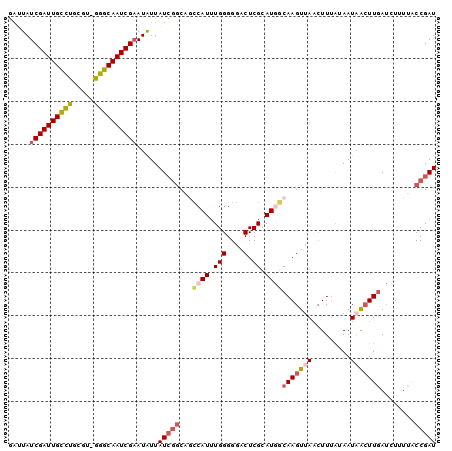
| Location | 10,953,561 – 10,953,660 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.54 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10953561 99 - 22224390 GCUUAUCGAUUGCUUGCGUCGGGCAAUCGAAUAUUAUCGGCAGCCAUAUGGGGGACUCGCAUU-CAAGUAAUAUUUAUAAUAAAUUGAGCUUUCGCCGAU .....(((((((((((...))))))))))).....((((((..((....))(..(((((....-...(((.....))).......)))).)..))))))) ( -26.94) >DroSec_CAF1 4199 100 - 1 GAAUAUCGAUUGCCUGCGUAGGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUGGAAUAACUUGAUAUUUUACCGAU .((((((((((((((.....))))))))).)))))(((((..(((((.(((....).)).)))))((((((.........)))))).........))))) ( -34.30) >DroSim_CAF1 1798 100 - 1 GAAUAUCGAUUGCCUGCGUAAGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUGGAAUAACUUCAUAUUUUACCGAU .((((((((((((((.....))))))))).)))))(((((..(((((.(((....).)).)))))((((((.........)))))).........))))) ( -34.30) >DroEre_CAF1 6756 99 - 1 GGUUAUCGAUUGUCUUUCU-GGGCAAUCGCAUGUUAUCACCAUUCAUUUGGGGGACUCGCAUAGCAAGUUAACUUUAUAAUCACUUGAUCUUUUACCGAU ((((((.((((((((....-))))))))((((((..((.(((......)))..))...)))).))...........)))))).................. ( -21.70) >DroYak_CAF1 3520 98 - 1 UAUUAUCGAUUGCCUUUCU-GGGCAAUCGAAUAUUAUAGGC-CCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUAAUAAUAGCUUGAUCUUUUACCGAU .....((((((((((....-))))))))))........(((-(((....)))).........(((((((((.........))))))).)).....))... ( -29.80) >consensus GAUUAUCGAUUGCCUGCGU_GGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUAUAAUAACUUGAUCUUUUACCGAU .....((((((((((.....)))))))))).....(((((...((((.(((....).)).))))(((((((.........)))))))........))))) (-21.10 = -22.54 + 1.44)

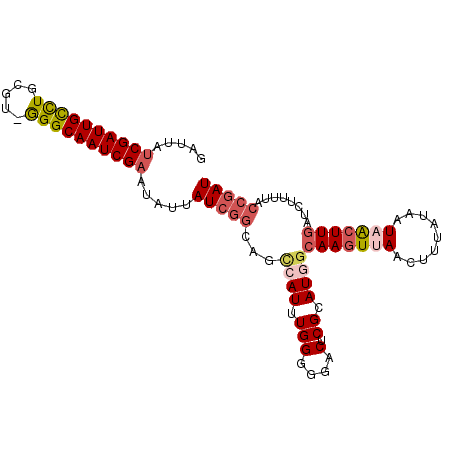
| Location | 10,953,581 – 10,953,682 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10953581 101 + 22224390 UUAUAAAUAUUACUUG-AAUGCGAGUCCCCCAUAUGGCUGCCGAUAAUAUUCGAUUGCCCGACGCAAGCAAUCGAUAAGCGCCGGCUAUUUUCGAUGGCCAA ...........(((((-....)))))........(((((..(((.(((..((((((((.........))))))))..(((....)))))).)))..))))). ( -26.40) >DroSec_CAF1 4219 102 + 1 UUCCAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCCUACGCAGGCAAUCGAUAUUCGCCGGCUAUUUUCGAUGGUCGA ...............(((((.(((.......(((((((((.(((......(((((((((.......)))))))))...))).)))))))))))))))))... ( -33.11) >DroSim_CAF1 1818 102 + 1 UUCCAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCUUACGCAGGCAAUCGAUAUUCGCCGGCUAUUUUCGAUGGCCGA ...............(((((.(((.......(((((((((.(((......((((((((((.....))))))))))...))).)))))))))))))))))... ( -36.51) >DroEre_CAF1 6776 101 + 1 UUAUAAAGUUAACUUGCUAUGCGAGUCCCCCAAAUGAAUGGUGAUAACAUGCGAUUGCCC-AGAAAGACAAUCGAUAACCGCCGGCUAUUUUCAAUGGCCCG ...........((((((...)))))).............((((........((((((.(.-.....).)))))).....))))((((((.....)))))).. ( -21.12) >DroYak_CAF1 3540 100 + 1 UUAUUAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGG-GCCUAUAAUAUUCGAUUGCCC-AGAAAGGCAAUCGAUAAUAGCCGGCUAUUUCCGAUGGCCCG ((((((.((((((((((...)))))).((((....)))-)....))))..(((((((((.-.....)))))))))))))))..(((((((...))))))).. ( -33.70) >consensus UUAUAAAGUUAACUUGCCAUGCGAGUCCCCCAAAUGGCUGCCGAUAAUAUUCGAUUGCCC_ACGCAGGCAAUCGAUAAUCGCCGGCUAUUUUCGAUGGCCCA ...........((((((...))))))...(((((((((((.(((......(((((((((.......)))))))))...))).)))))))).....))).... (-21.08 = -22.80 + 1.72)

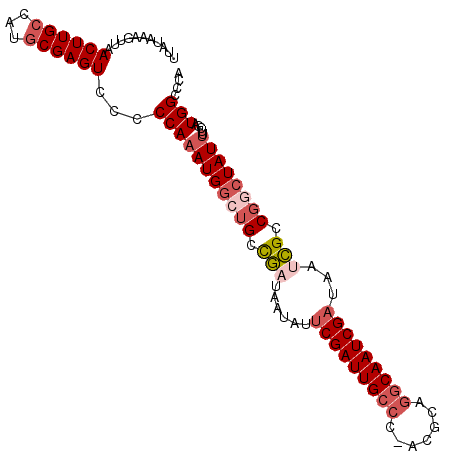

| Location | 10,953,581 – 10,953,682 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10953581 101 - 22224390 UUGGCCAUCGAAAAUAGCCGGCGCUUAUCGAUUGCUUGCGUCGGGCAAUCGAAUAUUAUCGGCAGCCAUAUGGGGGACUCGCAUU-CAAGUAAUAUUUAUAA .((((..((((.((((((....))...(((((((((((...))))))))))).)))).))))..))))...((....))......-................ ( -26.70) >DroSec_CAF1 4219 102 - 1 UCGACCAUCGAAAAUAGCCGGCGAAUAUCGAUUGCCUGCGUAGGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUGGAA ....(((..(......(((((..((((((((((((((.....))))))))).))))).))))).(((((.(((....).)).))))).......)..))).. ( -39.10) >DroSim_CAF1 1818 102 - 1 UCGGCCAUCGAAAAUAGCCGGCGAAUAUCGAUUGCCUGCGUAAGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUGGAA ....(((..(......(((((..((((((((((((((.....))))))))).))))).))))).(((((.(((....).)).))))).......)..))).. ( -39.40) >DroEre_CAF1 6776 101 - 1 CGGGCCAUUGAAAAUAGCCGGCGGUUAUCGAUUGUCUUUCU-GGGCAAUCGCAUGUUAUCACCAUUCAUUUGGGGGACUCGCAUAGCAAGUUAACUUUAUAA (.(((...(....)..))).).(((((.(((((((((....-))))))))((((((..((.(((......)))..))...)))).))..).)))))...... ( -24.80) >DroYak_CAF1 3540 100 - 1 CGGGCCAUCGGAAAUAGCCGGCUAUUAUCGAUUGCCUUUCU-GGGCAAUCGAAUAUUAUAGGC-CCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUAAUAA .(((((.((((......))))......((((((((((....-))))))))))........)))-)).(((.((..((((.((...)).))))..)).))).. ( -37.00) >consensus UCGGCCAUCGAAAAUAGCCGGCGAUUAUCGAUUGCCUGCGU_GGGCAAUCGAAUAUUAUCGGCAGCCAUUUGGGGGACUCGCAUGGCAAGUUAACUUUAUAA ...((((((((.....(((((......((((((((((.....))))))))))......))))).........(....)))).)))))............... (-25.46 = -26.26 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:09 2006