
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,948,880 – 10,948,979 |
| Length | 99 |
| Max. P | 0.893760 |

| Location | 10,948,880 – 10,948,979 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -18.61 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
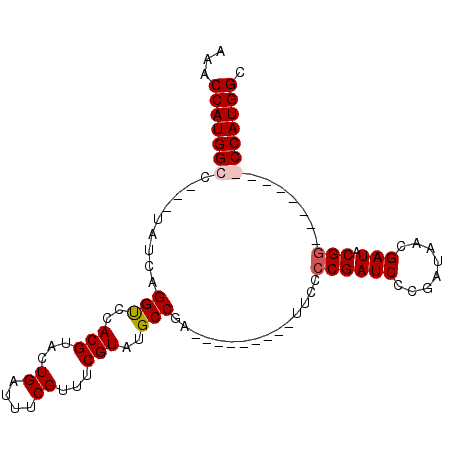
>X_DroMel_CAF1 10948880 99 + 22224390 AAACCAUGGCCCACUAUCAGGUCCACGUACUGAUUUCGUUUCGUGUGCCGAUUUGAC-GAUUUCCCGAUCCCGAUAACGAUACGGACGAUGGGCCAUGGC ...((((((((((...(((((((...(((((((.......))).)))).))))))).-......((((((........))).)))....)))))))))). ( -35.10) >DroSec_CAF1 6934 100 + 1 AAACCAUGGCAUACUAUCAGGUCCACGUACUGAUUUCGUUGCGUAUGCCGAUUUGACUGAUUUCCCGAUCCCGAUAACGAUACGGACGAUGGGCCAUGGC ...(((((((...(((((.(((..(((((.((....)).)))))..)))...............((((((........))).)))..)))))))))))). ( -31.00) >DroEre_CAF1 6811 80 + 1 AAACCAUGGCG---UAUCAGGCCCACGUACUGAUUUCGUUUCGUAUGCCGA---------UUCCGCGAUCCCGAUAACGAUACGG--------CCAUGGC ...((((((((---((((.(((..(((...((....))...)))..)))((---------((....))))........))))).)--------)))))). ( -26.60) >DroYak_CAF1 7432 80 + 1 AAACCAUGGCC---UAUCAGGUCCACGUACUGAUUUCGUUUCGUAUGCCGA---------UUCCCCGAUCCCGAUAACGAUACGG--------CCAUGGC ...((((((((---((((.(((..(((...((....))...)))..)))((---------((....))))........)))).))--------)))))). ( -26.10) >consensus AAACCAUGGCC___UAUCAGGUCCACGUACUGAUUUCGUUUCGUAUGCCGA_________UUCCCCGAUCCCGAUAACGAUACGG________CCAUGGC ...(((((((.........(((..(((...((....))...)))..)))...............((((((........))).))).......))))))). (-18.61 = -19.18 + 0.56)

| Location | 10,948,880 – 10,948,979 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10948880 99 - 22224390 GCCAUGGCCCAUCGUCCGUAUCGUUAUCGGGAUCGGGAAAUC-GUCAAAUCGGCACACGAAACGAAAUCAGUACGUGGACCUGAUAGUGGGCCAUGGUUU ((((((((((((..((((((....)).))))((((((.....-(((.....))).((((..((.......)).))))..)))))).)))))))))))).. ( -39.30) >DroSec_CAF1 6934 100 - 1 GCCAUGGCCCAUCGUCCGUAUCGUUAUCGGGAUCGGGAAAUCAGUCAAAUCGGCAUACGCAACGAAAUCAGUACGUGGACCUGAUAGUAUGCCAUGGUUU ((((((((......((((((....)).))))((((((......(((.....))).((((..((.......)).))))..)))))).....)))))))).. ( -28.00) >DroEre_CAF1 6811 80 - 1 GCCAUGG--------CCGUAUCGUUAUCGGGAUCGCGGAA---------UCGGCAUACGAAACGAAAUCAGUACGUGGGCCUGAUA---CGCCAUGGUUU (((((((--------(.((((((....)(((..((((...---------(((.....))).((.......)).))))..)))))))---))))))))).. ( -29.50) >DroYak_CAF1 7432 80 - 1 GCCAUGG--------CCGUAUCGUUAUCGGGAUCGGGGAA---------UCGGCAUACGAAACGAAAUCAGUACGUGGACCUGAUA---GGCCAUGGUUU (((((((--------((...(((((.(((.(.((((....---------)))))...)))))))).(((((.........))))).---))))))))).. ( -30.10) >consensus GCCAUGG________CCGUAUCGUUAUCGGGAUCGGGAAA_________UCGGCAUACGAAACGAAAUCAGUACGUGGACCUGAUA___GGCCAUGGUUU ((((((((.......(((..(((....)))...))).............((((..((((..((.......)).))))...))))......)))))))).. (-20.74 = -21.30 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:05 2006