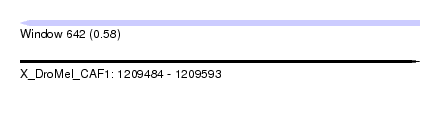
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,209,484 – 1,209,593 |
| Length | 109 |
| Max. P | 0.584392 |

| Location | 1,209,484 – 1,209,593 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -26.82 |
| Energy contribution | -25.43 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1209484 109 - 22224390 GUUGGGCUGCAUCAACGUGGCCGGCGUCAAGUGGGUCAUCAAGCUUCAGUUUAUACUGCUCAUGAUACUGCUGAUCUCGGCGCUGGACUUCAUGGUGGGCAGCUUCACU ((..((((((.(((.(((((((((((((.((..((((((..(((...(((....)))))).))))).)..))(....)))))))))...))))).)))))))))..)). ( -40.50) >DroVir_CAF1 3222 109 - 1 GCUCGGCUGCAUCAAUGUGGCUGGCGUCAAGUGGGUCAUUAAGCUGCAGUUUAUACUGCUGAUGGUAUUGCUUAUAUCGGCGUUAGAUUUCAUGGUUGGCAGCUUUGUG ((..((((((....(((...(((((((((((..(..(((((....(((((....))))))))))...)..))).....))))))))....))).....))))))..)). ( -34.90) >DroGri_CAF1 3808 109 - 1 GCUCGGCUGCAUUAAUGUGGCUGGUGUCAAGUGGGUGAUCAAAUUACAGUUUAUACUGCUCAUGAUUCUGCUCAUCUCAGCGCUGGAUUUCAUGGUCGGUAGCUUUAUU ....((((((....(((...(..((((..((((((((((((.....((((....))))....)))))..))))).))..))))..)....))).....))))))..... ( -30.40) >DroWil_CAF1 19880 109 - 1 UCUCGGCUGCAUCAAUGUGGCUGGCGUUAAAUGGGUAAUCAAAUUGCAAUUCAUAUUGCUAAUGAUUUUACUAAUAUCGGCGCUGGACUUUAUGGUGGGCAGCUUUAUA ....((((((....(((.(((..((((..(...(((((....((((((((....))))).)))....)))))....)..))))..).)).))).....))))))..... ( -31.30) >DroYak_CAF1 1857 109 - 1 ACUAGGCUGCAUUAACGUGGCCGGCGUCAAGUGGGUCAUUAAGCUUCAGUUUAUACUGCUCAUGAUUUUGCUGAUCUCGGCGCUGGACUUCAUGGUGGGCAGCUUCACU ...(((((((.....(((((((((((((..((((((...(((((....)))))....))))))((((.....))))..))))))))...)))))....))))))).... ( -38.80) >DroMoj_CAF1 1711 109 - 1 GCUCGGCUGCAUCAAUGUGGCAGGCGUCAAGUGGGUGAUCAAACUGCAGUUUAUUCUGCUGAUGAUAUUGCUCAUAUCUGCGCUGGAUUUCAUGGUAGGCAGCUUUGUG ((..((((((....(((...(.(((((...((((((.((((....((((......))))...))))...))))))....))))).)....))).....))))))..)). ( -33.90) >consensus GCUCGGCUGCAUCAAUGUGGCUGGCGUCAAGUGGGUCAUCAAACUGCAGUUUAUACUGCUCAUGAUAUUGCUCAUAUCGGCGCUGGACUUCAUGGUGGGCAGCUUUAUU ....((((((.....((((.((((((((..((.(((.((((.....((((....))))....))))...))).))...))))))))....))))....))))))..... (-26.82 = -25.43 + -1.38)

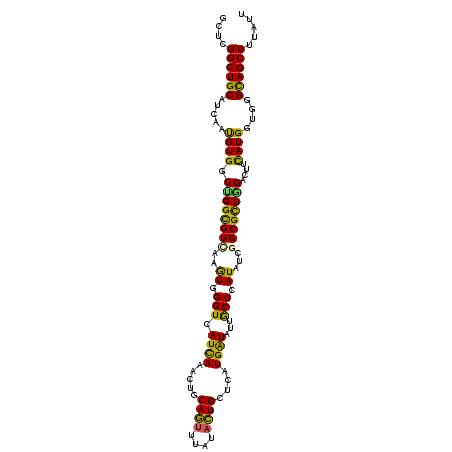
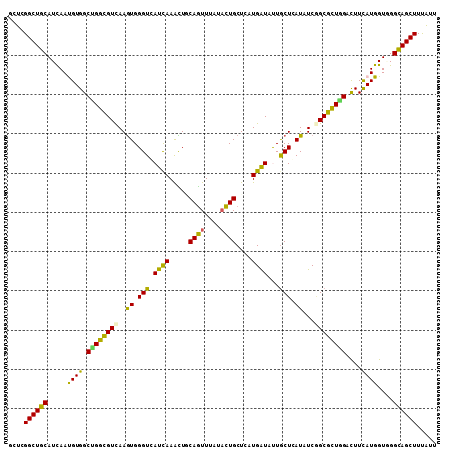
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:06 2006