| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,924,637 – 10,924,727 |
| Length | 90 |
| Max. P | 0.834967 |

| Location | 10,924,637 – 10,924,727 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 56.68 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -9.36 |
| Energy contribution | -10.75 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10924637 90 + 22224390 ----------------GAGCCCAGGACGACAAGAUCGAAAACAA------------GCAAGAGGGAGCUCAUGAAGAUCCUCUUCGAUGGCUUCCUCAGUUGCUUGGAGAAGGAGAAC ----------------....((....(((.....)))....(((------------((((((((((((.(((((((.....)))).)))))))))))..))))))).....))..... ( -31.90) >DroSec_CAF1 7724 72 + 1 ----------------GAGCCCAGG------------------A------------GCAAGAGGGAGCUCAUGAAGAUCCUCUUCGAUGGCUUCCUCAGUUGCUUGGAGAAGGAGAAC ----------------....(((((------------------(------------((..((((((((.(((((((.....)))).))))))))))).))).)))))........... ( -27.80) >DroSim_CAF1 7161 72 + 1 ----------------GAGCCCAGG------------------A------------GCAAGAGGGAGCUCAUGAAGAUCCUCUUCGAUGGCUUCCUCAGUUGCNNNNNNNNNNNNNNN ----------------.........------------------.------------((((((((((((.(((((((.....)))).)))))))))))..))))............... ( -22.30) >DroEre_CAF1 7588 79 + 1 ----------------GCACCCAAGU---GCAGACCGAACACAA------------CCAAGAGGGUGCUCAUCAGGAUCCGCUUCGAUGGCUUCCUCGGCUGCUUGGAUA-------- ----------------....((((..---((((.(((.......------------.)..(((((.((.(((((((.....))).)))))).)))))))))))))))...-------- ( -25.50) >DroYak_CAF1 7725 99 + 1 ----------------GAGUCCAAGU---GCAGACCGAAAACAACAACAACAGCAACAAAGAGGGUGCUCAUGAGGAUCCUCUUCGAUGGCUUCCUCAGUUGCUUGGAGAAGGAGAAC ----------------...(((((((---...........))..........(((((...(((((.((.(((((((.....)))).))))).))))).)))))))))).......... ( -25.40) >DroMoj_CAF1 11096 97 + 1 CGGGAUCAAGUACCUCGCCUCGAAGU---GCAGGUCGAGUCGAU------------UUAUGGGCUUGCAC------CUGAUGUUCAAUGGGUUCUUGAGCUGCCUGGAGCGGGAGUAC .........(((((((((..((((..---.(((((((((((...------------.....)))))).))------)))...)))...((((.........)))))..))))).)))) ( -32.90) >consensus ________________GAGCCCAAGU___GCAGA_CGAA_ACAA____________GCAAGAGGGAGCUCAUGAAGAUCCUCUUCGAUGGCUUCCUCAGUUGCUUGGAGAAGGAGAAC ............................................................(((((.((.(((((((.....)))).))))).)))))..................... ( -9.36 = -10.75 + 1.39)

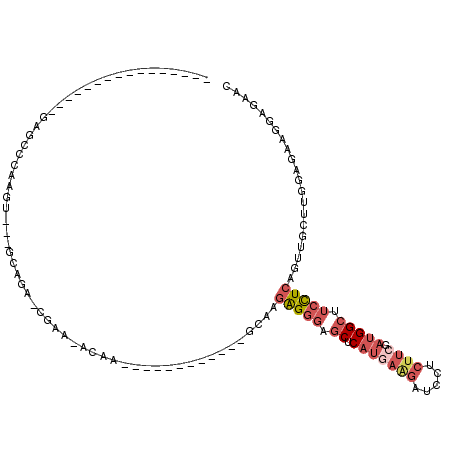

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:57 2006