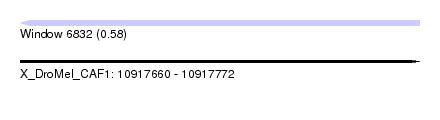
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,917,660 – 10,917,772 |
| Length | 112 |
| Max. P | 0.575587 |

| Location | 10,917,660 – 10,917,772 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -41.35 |
| Consensus MFE | -22.35 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
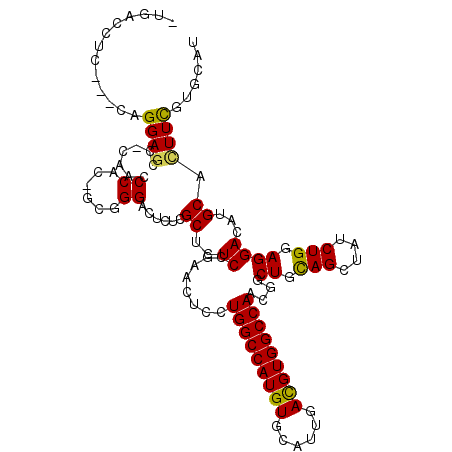
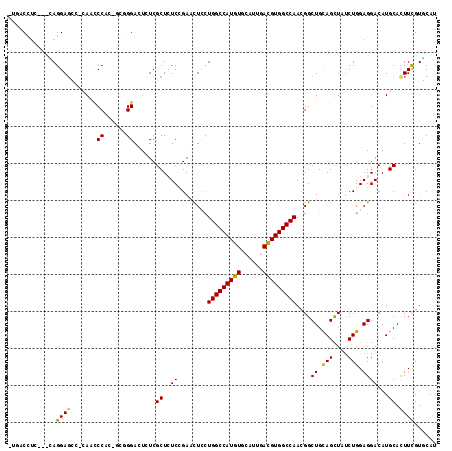
>X_DroMel_CAF1 10917660 112 - 22224390 -UGCCCUC---UAGGAGCCACAACCCACUGCUGGACUCUCGCUCUCCGAGCUCCUGGCCAUGUGCAUUGAUGUGGCCAACGGCUGCAGUUAUCUGGAGGACAUGCACUUUGUGCAU -...((((---(((((((......(((....)))......))))....((((..(((((((((......)))))))))..))))........)))))))..((((((...)))))) ( -40.20) >DroPse_CAF1 723 106 - 1 -AAUCAUG---AAGGAGG--UAGUCCA----GGGACUGUCGCUGUCGGAAUUGCUGGCCAUGUGCAUUGACGUGGCCAACGGCUGCAGCUAUCUGGAGGACAUGCACUUCGUUCAU -....(((---((.((((--(..((((----((..((((.(((((.........(((((((((......)))))))))))))).))))...))))))(......)))))).))))) ( -41.80) >DroEre_CAF1 711 112 - 1 -UGACCUC---CAGGAGCCGCAACCCACCGCUGGACUCUCGCUCUCUGAACUCCUGGCCAUGUGCAUUGAUGUGGCCAACGGCUGCAGUUACCUGGAGGACAUGCACUUCGUGCAC -...((((---(((((((.(((.((.......(((...(((.....)))..)))(((((((((......)))))))))..)).))).))).))))))))...(((((...))))). ( -44.10) >DroYak_CAF1 743 115 - 1 -UGCCCAUCAGCAGGAGUCGCAACCCGCAGCUGGACUCUCGCUCUCCGAGCUCCUGGCCAUGUGCAUUGAUGUGGCCAAUGGCUGUAGUUAUCUGGAGGACAUGCAUUUCGUGCAC -.........(((.(((..(((....(((((((((..((((.....)))).)))(((((((((......)))))))))..)))))).(((........))).)))..))).))).. ( -38.80) >DroAna_CAF1 731 113 - 1 CUUACUUU---CAGGACUCCCAGCCCGGUCAGGGAUUGUCGCUCUCCGAGCUCUUGGCCAUGUGCCUGGACGUGGCCAAUGGCUGUAGCUAUCUGGAGGACAUGCACUUUGUCCAU ......((---((((((......(((.....)))...)))(((...(.((((.((((((((((......)))))))))).))))).)))...)))))(((((.......))))).. ( -42.00) >DroPer_CAF1 723 106 - 1 -AAUCUUG---AAGGAGG--UAGUCCA----GGGACUGUCGCUGUCGGAAUUGCUGGCCAUGUGCAUUGACGUGGCCAACGGCUGCAGCUAUCUGGAGGACAUGCACUUCGUUCAU -.....((---((.((((--(..((((----((..((((.(((((.........(((((((((......)))))))))))))).))))...))))))(......)))))).)))). ( -41.20) >consensus _UGACCUC___CAGGAGCC_CAACCCAC_GCGGGACUCUCGCUCUCCGAACUCCUGGCCAUGUGCAUUGACGUGGCCAACGGCUGCAGCUAUCUGGAGGACAUGCACUUCGUGCAU .............((((.......((......))......((..((........(((((((((......)))))))))....((.(((....))).))))...)).))))...... (-22.35 = -21.72 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:52 2006