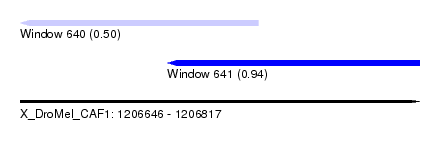
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,206,646 – 1,206,817 |
| Length | 171 |
| Max. P | 0.944101 |

| Location | 1,206,646 – 1,206,748 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -8.63 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
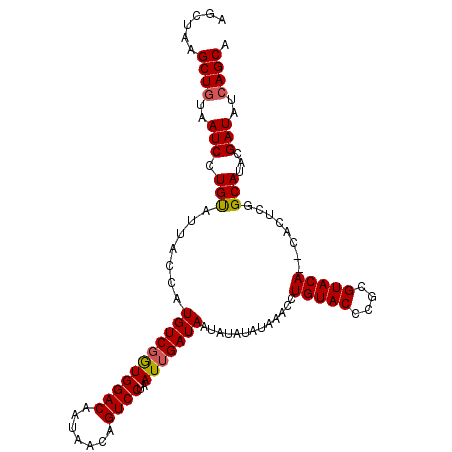
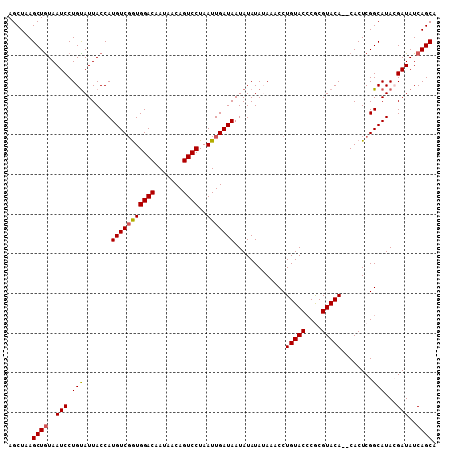
>X_DroMel_CAF1 1206646 102 - 22224390 CUGUACCCGCGUACACUCACUCGGCAUA-CGAUAUCAGCAAUAACCGCACUC--------UAUAUACACCCAAUAU-------AUGUAUAUAUAUCACUCGCAUAUAUCC-GAUAUAUA .((((.(((.((......)).)))..))-))(((((.((.......))....--------((((((((........-------.))))))))..................-)))))... ( -14.20) >DroSec_CAF1 16297 100 - 1 CUGUACACGCGUACA--CACUCGGCAUA-CGAUAUCAGCAAUAACCGCACUC--------UAUAUACACCCAAUAU-------AUGUAUAUAUAUCACUCGCAUAUAUCC-GAUAUAUA .(((((....)))))--...((((.(((-(((.....((.......))....--------((((((((........-------.))))))))......)))....)))))-))...... ( -14.60) >DroSim_CAF1 8883 100 - 1 CUGUACCCGCGUACA--CACUCGGCAUA-CGAUAUCAGCAAUAACCGCACUC--------UAUAUACACCCAAUAU-------AUGUAUAUAUAUCACUCGCAUAUAUCC-GAUAUAUA .(((((....)))))--...((((.(((-(((.....((.......))....--------((((((((........-------.))))))))......)))....)))))-))...... ( -14.00) >DroEre_CAF1 16967 100 - 1 CUGUACCCACGUACA--CACUCGGCAUA-AGAUAUCAGCAAUAACCGCACUC--------UAUAUACACCCAAUAU-------AUGUAUAUAUAUCACUCGCAUAUAUCC-GAUAUAUA .(((((....)))))--...((((.(((-........((.......))....--------((((((((........-------.)))))))).............)))))-))...... ( -13.70) >DroYak_CAF1 20971 98 - 1 CUGUACCCACGUACA--CACUCGGCAUA-AGAUAUCAGCAAUAACCGCAC----------UAUAUACACCCAAUAU-------AUGUAUAUAUAUCAGUCGCAUAUAUCC-GAUAUAUA .(((((....)))))--...((((.(((-.(((....((.......))..----------((((((((........-------.)))))))).....))).....)))))-))...... ( -13.80) >DroAna_CAF1 15323 109 - 1 CAGCAGCCAU--ACA--UA-----CAUACAUAUAUCAGCAAUAACCGCACCCAAACAUCCUGUAU-UACCCUAUAUUAAUGUAAUGUAUAUACAAUGUAUACUUAUACCGGGAUAUAUA ..........--...--..-----.....(((((((.((.......))..((..((((...((((-......))))..))))...((((((.....)))))).......))))))))). ( -14.60) >consensus CUGUACCCACGUACA__CACUCGGCAUA_AGAUAUCAGCAAUAACCGCACUC________UAUAUACACCCAAUAU_______AUGUAUAUAUAUCACUCGCAUAUAUCC_GAUAUAUA ..((((....)))).................(((((.((.......))............((((((((................))))))))...................)))))... ( -8.63 = -9.02 + 0.39)
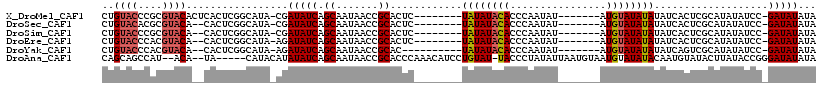
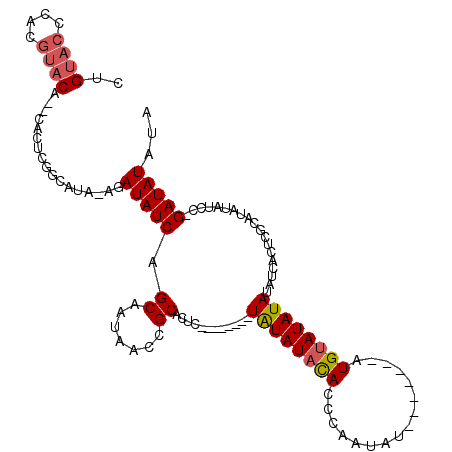
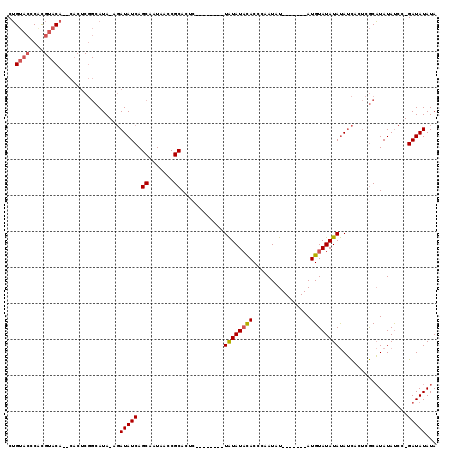
| Location | 1,206,709 – 1,206,817 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1206709 108 - 22224390 AGCUAAGCUAUAAUCCUGUAUUACCAUGUCGGUGGACAAUAACAGUCCUAAUUGAUAAUAUAUAUAAACCUGUACCCGCGUACACUCACUCGGCAUACGAUAUCAGCA ......(((...(((.(((((.....(((((((((((.......))))..)))))))....))))).....(((.(((.((......)).)))..))))))...))). ( -18.90) >DroSec_CAF1 16360 106 - 1 AGCUAAGCUGUAAUCCUGUAUUACCAUGUCUGUGGACAAUAACAGUCCUAAUUGAUAAUAUAUAUAAACCUGUACACGCGUACA--CACUCGGCAUACGAUAUCAGCA ......((((..(((..((((..((.((((...((((.......)))).....)))).............(((((....)))))--.....)).)))))))..)))). ( -20.90) >DroSim_CAF1 8946 106 - 1 AGCUAAGCUGUAAUCCUGUAUUACCAUGUCGGUGGACAAUAACAGUCCUAAUUGAUAAUAUAUAUAAACCUGUACCCGCGUACA--CACUCGGCAUACGAUAUCAGCA ......((((..(((..((((..((.(((((((((((.......))))..))))))).............(((((....)))))--.....)).)))))))..)))). ( -22.20) >DroEre_CAF1 17030 105 - 1 AGCUAAGCUGUAAUCCUGCAUUACCAUGUCGAUGGACAAUAACAGUCCUAAUUGAUAAUAUAUA-AAACCUGUACCCACGUACA--CACUCGGCAUAAGAUAUCAGCA ......((((..(((.(((.......(((((((((((.......))))..))))))).......-.....(((((....)))))--......)))...)))..)))). ( -22.70) >DroYak_CAF1 21032 105 - 1 AGCUAAGCUGUAAUCCUGUAUUACCAUGUCGGUGGACAAUAACAGUCCUAAUUGAUAAUAUAUA-AAACCUGUACCCACGUACA--CACUCGGCAUAAGAUAUCAGCA ......((((..(((.(((....((.(((((((((((.......))))..))))))).......-.....(((((....)))))--.....)).))).)))..)))). ( -21.00) >consensus AGCUAAGCUGUAAUCCUGUAUUACCAUGUCGGUGGACAAUAACAGUCCUAAUUGAUAAUAUAUAUAAACCUGUACCCGCGUACA__CACUCGGCAUACGAUAUCAGCA ......((((..(((.(((.......(((((((((((.......))))..))))))).............(((((....)))))........)))...)))..)))). (-19.28 = -19.36 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:05 2006