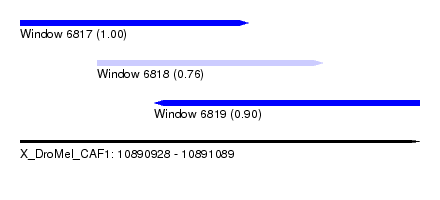
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,890,928 – 10,891,089 |
| Length | 161 |
| Max. P | 0.996222 |

| Location | 10,890,928 – 10,891,020 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.22 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
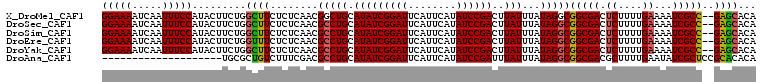
>X_DroMel_CAF1 10890928 92 + 22224390 -----GGUGGUGUGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA -----.((((.((((.((((((((((...((((((((........)))))))))-------))))))).))))--((((((((......))))))))))))))... ( -34.60) >DroSec_CAF1 44919 92 + 1 -----GGUGGUGUGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA -----.((((.((((.((((((((((...((((((((........)))))))))-------))))))).))))--((((((((......))))))))))))))... ( -34.60) >DroSim_CAF1 47901 92 + 1 -----GGUGGUGUGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGGAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA -----.((((.((((.((((((((((...((((((((........)))))))))-------))))))).))))--((((((((......))))))))))))))... ( -34.20) >DroEre_CAF1 21890 92 + 1 -----GGUGGUGCGAUGCCACAUUGUUAUGUUUUGCUUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA -----.((((.((((.((((((((((...((((((............)))))))-------))))))).))))--((((((((......))))))))))))))... ( -33.40) >DroYak_CAF1 45624 92 + 1 -----GGUGGUGCGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA -----.((((.((((.((((((((((...((((((((........)))))))))-------))))))).))))--((((((((......))))))))))))))... ( -37.30) >DroAna_CAF1 55870 106 + 1 GGUUGGGCGGUUCUCUGCCGCAUUGUUGUGUUUUGUUUUAUUUAUAACGGAGCGGGACGCUCGAUGUGUGUGCGGAGCGAUAUUCAAAAGCGUCGCCGCCUAUAAA ...((((((((.(((((((((((((..(((((((((((..(.....)..))))))))))).)))))))...)))))).(((..........))))))))))).... ( -39.70) >consensus _____GGUGGUGCGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG_______CAAUGUGUGCUC__GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAA .....(((........(((((((((...(((((((((........))))))))).......))))))).))....((((((((......)))))))))))...... (-29.63 = -29.22 + -0.41)

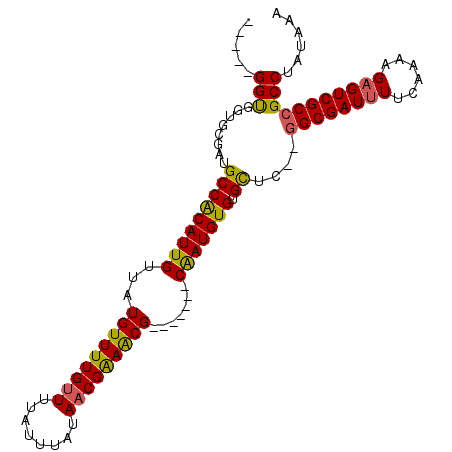
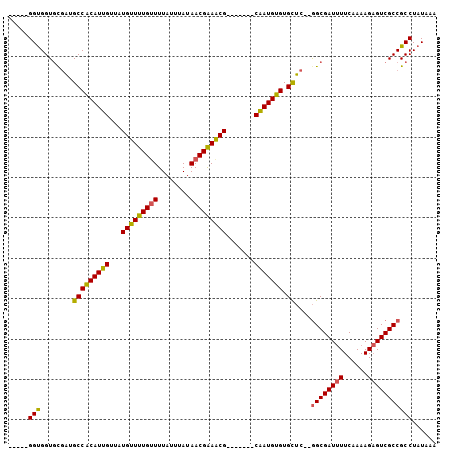
| Location | 10,890,959 – 10,891,050 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10890959 91 + 22224390 UUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((..((.(((-------(...))))..))--((((((((......))))))))...)))))))))(((((((........)))))))..... ( -28.20) >DroSec_CAF1 44950 91 + 1 UUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((..((.(((-------(...))))..))--((((((((......))))))))...)))))))))(((((((........)))))))..... ( -28.20) >DroSim_CAF1 47932 91 + 1 UUUAUUUAUAACGGAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((..((...(-------((.....))).(--((((((((......))))))))))))))))))))(((((((........)))))))..... ( -29.00) >DroEre_CAF1 21921 91 + 1 UUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((..((.(((-------(...))))..))--((((((((......))))))))...)))))))))(((((((........)))))))..... ( -28.20) >DroYak_CAF1 45655 91 + 1 UUUAUUUAUAACGAAACG-------CAAUGUGUGCUC--GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((..((.(((-------(...))))..))--((((((((......))))))))...)))))))))(((((((........)))))))..... ( -28.20) >DroAna_CAF1 55906 100 + 1 UUUAUUUAUAACGGAGCGGGACGCUCGAUGUGUGUGCGGAGCGAUAUUCAAAAGCGUCGCCGCCUAUAAAUAAAUCGGAUAUGAAUGAAUCCGAUAUGCA .((((((((...((.((((((((((...((.((((((...)).)))).))..)))))).))))))))))))))(((((((........)))))))..... ( -31.70) >consensus UUUAUUUAUAACGAAACG_______CAAUGUGUGCUC__GGCGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAUGAAUGAAUCCGAUAUGCA .(((((((((.....(((..........)))........((((((((......))))))))...)))))))))(((((((........)))))))..... (-21.66 = -21.72 + 0.06)

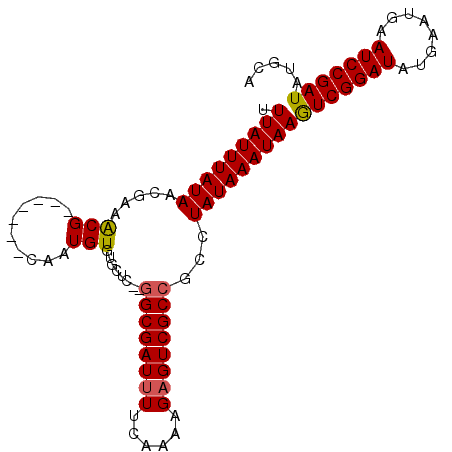

| Location | 10,890,982 – 10,891,089 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10890982 107 - 22224390 GGAAAAUCAAUUUCCAUACUUCUGGCUUCUCUCAACGGCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC--GAGCACA (((((.....)))))........((((.........))))((...((((((........))))))............((((((.((....))...)))))--).))... ( -25.00) >DroSec_CAF1 44973 107 - 1 GGAAAAUCAAUUUCCAUACUUCUGGCUUCUCUCAACGCCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC--GAGCACA (((((.....))))).........((((........(((((.(((((((((........))))))..)))...)))))(((((.((....))...)))))--))))... ( -29.00) >DroSim_CAF1 47955 107 - 1 GGAAAAUCAAUUUCCAUACUUCUGGCUUCUCUCAACGCCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC--GAGCACA (((((.....))))).........((((........(((((.(((((((((........))))))..)))...)))))(((((.((....))...)))))--))))... ( -29.00) >DroEre_CAF1 21944 107 - 1 GGAAAAUCAAUUUCCAUACUUCUGGUUUCUCUCAACGCCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC--GAGCACA (((((.....)))))...............(((...(((((.(((((((((........))))))..)))...)))))(((((.((....))...)))))--))).... ( -27.60) >DroYak_CAF1 45678 107 - 1 GGAAAAUCAAUUUCCAUACUUCUGGCUUCUCUCAACGCCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC--GAGCACA (((((.....))))).........((((........(((((.(((((((((........))))))..)))...)))))(((((.((....))...)))))--))))... ( -29.00) >DroAna_CAF1 55936 89 - 1 --------------------UGCGCUGUCUUUCGACGCCUGCAUAUCGGAUUCAUUCAUAUCCGAUUUAUUUAUAGGCGGCGACGCUUUUGAAUAUCGCUCCGCACACA --------------------(((((.(((....)))))..((..(((((((........))))))).((((((.(((((....))))).))))))..))...))).... ( -27.30) >consensus GGAAAAUCAAUUUCCAUACUUCUGGCUUCUCUCAACGCCUGCAUAUCGGAUUCAUUCAUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCGCC__GAGCACA (((((.....))))).........((((........(((((.(((((((((........))))))..)))...)))))(((((.((....))...)))))..))))... (-22.50 = -22.43 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:39 2006