
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,873,658 – 10,873,769 |
| Length | 111 |
| Max. P | 0.974298 |

| Location | 10,873,658 – 10,873,769 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.14 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -33.02 |
| Energy contribution | -33.78 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10873658 111 + 22224390 -UCAUCUUGGCCAGCCACGUCACGCAGCUGGUGAAAAUUUGCUGGCCAGUUGGCCAUCGGGACAACCGAGCAUCAGAUGGCUGACUGGGCAUAUUUUU-UUUCCCCCAACAGC -......((((((((....((((.......))))......))))))))((..((((((((.....))(.....).))))))..))((((.........-.....))))..... ( -39.74) >DroSec_CAF1 28408 113 + 1 AUCAUCUUGGCCAGCCACGUCACGCAGCUGGUGAAAAUUUGCUGGCCCGUUGGGCAUCGGGACAACCGAGCAUCAGAUGGCUGACUGGGCAUAUUUUUUUUUCGCCCAACAGC ........(((((((....((((.......))))......))))))).(((((((.((((.....))))((.((((........)))))).............)))))))... ( -42.60) >DroSim_CAF1 29808 112 + 1 -UCAUCUUGGCCAGCCACGUCACGCAGCUGGUGAAAAUUUGCUGGCCAGUUGGCCAUCGGGACAACCGAGCAUCAGAUGGCUGACUGGGCAUAUUUUUUUUUCGCCCAACAGC -......((((((((....((((.......))))......))))))))((..((((((((.....))(.....).))))))..))(((((.............)))))..... ( -44.02) >DroEre_CAF1 4575 107 + 1 -UCAUCUUGGCCAGCCACGUCACGCAGAUGGUGAAAAUUUGCUGGCCAGUUGGCCAACGGGACAUUCGAGCAUCAGAUGGCUGACGGGGCAUAUUU-UUU----CCCAGCGGC -.(((((((((((((...((((.((((((.......))))))))))..)))))))).(((.....)))......)))))((((..((((.......-..)----)))..)))) ( -36.50) >DroYak_CAF1 27976 108 + 1 -UCAUCUUGGCCAGCCACGUCACGCAGAUGGUGAAAAUUUGCAGGCCAGUUGGCCAACGGGACAUUCGAGCAUCAGAUGGCUGACUGGGCAUAUUUUUUU----CCCAACGGC -.(((((((((((((...(((..((((((.......)))))).)))..)))))))).(((.....)))......)))))((((..((((.(........)----)))).)))) ( -32.30) >consensus _UCAUCUUGGCCAGCCACGUCACGCAGCUGGUGAAAAUUUGCUGGCCAGUUGGCCAUCGGGACAACCGAGCAUCAGAUGGCUGACUGGGCAUAUUUUUUUUUC_CCCAACAGC .......((((((((....((((.......))))......))))))))((..((((((((.....))(.....).))))))..)).(((...............)))...... (-33.02 = -33.78 + 0.76)

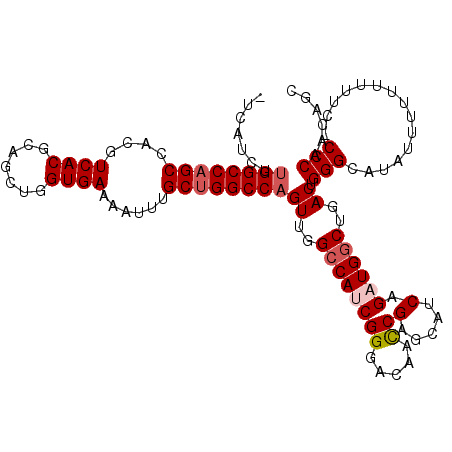

| Location | 10,873,658 – 10,873,769 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.14 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -32.78 |
| Energy contribution | -34.42 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10873658 111 - 22224390 GCUGUUGGGGGAAA-AAAAAUAUGCCCAGUCAGCCAUCUGAUGCUCGGUUGUCCCGAUGGCCAACUGGCCAGCAAAUUUUCACCAGCUGCGUGACGUGGCUGGCCAAGAUGA- (((((((((.....-.........))))).))))(((((..(((((((.....))))(((((....))))))))........(((((..((...))..)))))...))))).- ( -44.34) >DroSec_CAF1 28408 113 - 1 GCUGUUGGGCGAAAAAAAAAUAUGCCCAGUCAGCCAUCUGAUGCUCGGUUGUCCCGAUGCCCAACGGGCCAGCAAAUUUUCACCAGCUGCGUGACGUGGCUGGCCAAGAUGAU (((((((((((...........))))))).))))(((((((......((((.((((........)))).))))......)).(((((..((...))..)))))...))))).. ( -44.10) >DroSim_CAF1 29808 112 - 1 GCUGUUGGGCGAAAAAAAAAUAUGCCCAGUCAGCCAUCUGAUGCUCGGUUGUCCCGAUGGCCAACUGGCCAGCAAAUUUUCACCAGCUGCGUGACGUGGCUGGCCAAGAUGA- (((((((((((...........))))))).))))(((((..(((((((.....))))(((((....))))))))........(((((..((...))..)))))...))))).- ( -47.80) >DroEre_CAF1 4575 107 - 1 GCCGCUGGG----AAA-AAAUAUGCCCCGUCAGCCAUCUGAUGCUCGAAUGUCCCGUUGGCCAACUGGCCAGCAAAUUUUCACCAUCUGCGUGACGUGGCUGGCCAAGAUGA- ((((.((((----(..-..((.((...((((((....))))))..)).)).))))).))))((.(((((((((......(((((....).))))....))))))).)).)).- ( -37.60) >DroYak_CAF1 27976 108 - 1 GCCGUUGGG----AAAAAAAUAUGCCCAGUCAGCCAUCUGAUGCUCGAAUGUCCCGUUGGCCAACUGGCCUGCAAAUUUUCACCAUCUGCGUGACGUGGCUGGCCAAGAUGA- ..(((((((----...........))).(((((((((.(.((((..(.....)..((.((((....)))).))...............)))).).)))))))))...)))).- ( -32.50) >consensus GCUGUUGGG_GAAAAAAAAAUAUGCCCAGUCAGCCAUCUGAUGCUCGGUUGUCCCGAUGGCCAACUGGCCAGCAAAUUUUCACCAGCUGCGUGACGUGGCUGGCCAAGAUGA_ (((((((((...............))))).))))(((((..(((.(((.....))).(((((....))))))))........(((((..((...))..)))))...))))).. (-32.78 = -34.42 + 1.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:29 2006