
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,870,721 – 10,870,891 |
| Length | 170 |
| Max. P | 0.869831 |

| Location | 10,870,721 – 10,870,832 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -36.76 |
| Energy contribution | -37.56 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
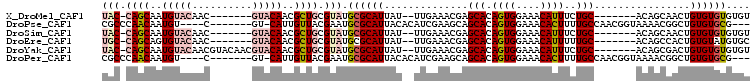
>X_DroMel_CAF1 10870721 111 - 22224390 UACGCAGCGUUGUAC-------GUUGUACAUUGCUGGUAAAAACGCGGCCAACGUCAAAGGCAAAUGGCUCAAGUGGUGGGCGUUUGACAAGAGCGGGCGUGGCAGUGGGCGGCGGGG- (((.(((((.(((((-------...))))).))))))))....(((.(((.((((((((.((..((.((....)).))..)).))))))....((.......)).)).))).)))...- ( -43.10) >DroSec_CAF1 25295 111 - 1 UACGCAGCGUUGUAC-------GUUGUACAUUGCUGGUAAAAACGCGACCAACGUCAAAGGCAAAUGGCUCAAGUGGUGGGCGUUUGACAAGAGCGGGCGUGGCAGUGGGCGGCGGGG- (((.(((((.(((((-------...))))).))))))))....(.((.((...((((((.((..((.((....)).))..)).))))))....((..((......))..)))))).).- ( -37.70) >DroSim_CAF1 26714 111 - 1 UACGCAGCGUUGUAC-------GUUGUACAUUGCUGGUAAAAACGCGGCCAACGUCAAAGGCAAAUGGCUCAAGUGGUGGGCGUUUGACAAGAGCGGGCGUGGCAGCGGGCGGCGGGG- (((.(((((.(((((-------...))))).))))))))....(((.(((...((((((.((..((.((....)).))..)).))))))....((..((...)).)).))).)))...- ( -43.40) >DroEre_CAF1 1725 111 - 1 UACGCAGCGUUGUAC-------GUUGUACACUGCUGGCAAAAACGCGGCCAACGUCAAAGGCAAAUGGCUCAAGUGGUGGGCGUUCGACAAGAGCGGGCGUGGCAGGGGGCGGUGGGG- ...((((((.....)-------))))).(((((((.((......)).((((.((((...........(((((.....)))))((((.....)))).))))))))....)))))))...- ( -41.20) >DroYak_CAF1 25184 119 - 1 UACGCAGCGUUGUACGUUGUACGUUGUACAUUGCUGGUAAAAACGCGGCCAACGUCAAAGGCAAACGGCUCAAGUGGAGGGCGUUUGACAAGAGCGGGCGUGGCAGUGGGCGGUGGGGG (((.(((((.((((((........)))))).))))))))....(((.(((.((((((((.((...(.((....)).)...)).))))))....((.......)).)).))).))).... ( -42.10) >consensus UACGCAGCGUUGUAC_______GUUGUACAUUGCUGGUAAAAACGCGGCCAACGUCAAAGGCAAAUGGCUCAAGUGGUGGGCGUUUGACAAGAGCGGGCGUGGCAGUGGGCGGCGGGG_ (((.(((((.(((((..........))))).))))))))....(((.(((.((((((((.((..((.((....)).))..)).))))))....((.......)).)).))).))).... (-36.76 = -37.56 + 0.80)

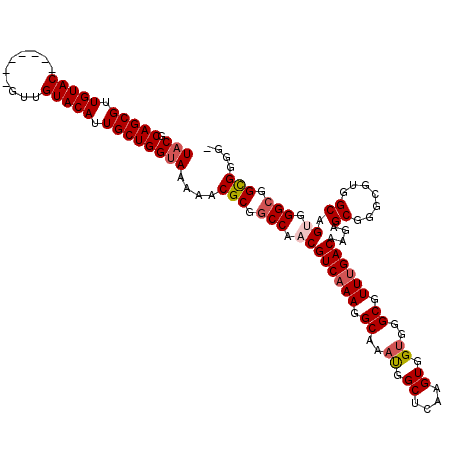

| Location | 10,870,760 – 10,870,870 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -21.15 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10870760 110 + 22224390 CACCACUUGAGCCAUUUGCCUUUGACGUUGGCCGCGUUUUUAC-CAGCAAUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCU ..(((((.(.(((....(((.........))).((((...(((-((((..(((((...-------)))))..)))).))).)))).....--.......).)).)))))).......... ( -30.50) >DroSec_CAF1 25334 110 + 1 CACCACUUGAGCCAUUUGCCUUUGACGUUGGUCGCGUUUUUAC-CAGCAAUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCU ..(((((.(.(((..........(((....)))((((...(((-((((..(((((...-------)))))..)))).))).)))).....--.......).)).)))))).......... ( -29.70) >DroSim_CAF1 26753 110 + 1 CACCACUUGAGCCAUUUGCCUUUGACGUUGGCCGCGUUUUUAC-CAGCAAUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCU ..(((((.(.(((....(((.........))).((((...(((-((((..(((((...-------)))))..)))).))).)))).....--.......).)).)))))).......... ( -30.50) >DroEre_CAF1 1764 110 + 1 CACCACUUGAGCCAUUUGCCUUUGACGUUGGCCGCGUUUUUGC-CAGCAGUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUUU ..(((((.(.(((....(((.........))).((((...(((-((((..(((((...-------)))))..)))).))).)))).....--.......).)).)))))).......... ( -31.80) >DroYak_CAF1 25224 117 + 1 CUCCACUUGAGCCGUUUGCCUUUGACGUUGGCCGCGUUUUUAC-CAGCAAUGUACAACGUACAACGUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCU .((((((.(.((((((((((.........))).((((...(((-((((..(((((..........)))))..)))).))).)))).....--...))))).)).)))))))......... ( -37.40) >DroPer_CAF1 47133 88 + 1 CAGC--------------------ACGGUGGCCGUGCUUUCGCCCAACAAUGU----C-------GU-CAUUGUUACGAAUGCGCAUUACACAUCGAAGCAGCACAGUGGAAACACUUUU ..((--------------------.(((((...((((.((((...(((((((.----.-------..-))))))).)))).))))......)))))..)).....((((....))))... ( -27.40) >consensus CACCACUUGAGCCAUUUGCCUUUGACGUUGGCCGCGUUUUUAC_CAGCAAUGUACAAC_______GUACAACGCUGCGUAUGCGCAUUAU__UUGAAACGAGCACAGUGGAAACAUUUCU .....((((.((((..............)))).((((...(((.((((..(((((..........)))))..)))).))).)))).............))))...((((....))))... (-21.15 = -22.46 + 1.31)
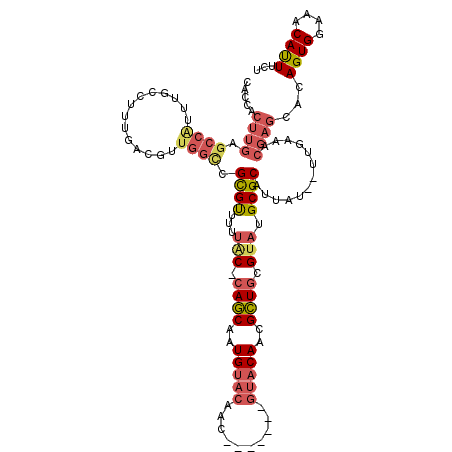
| Location | 10,870,800 – 10,870,891 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.59 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10870800 91 + 22224390 UAC-CAGCAAUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCUGC-------ACAGCAACUGUGUGUGUGU (((-((((..(((((...-------)))))..)))).))).((((((...--.........(((.((((....))))..)))-------((((...)))))))))).. ( -25.00) >DroPse_CAF1 46896 93 + 1 CGCCCAACAAUGU----C-------GU-CAUUGUUACGAAUGCGCAUUACACAUCGAAGCAGCACAGUGGAAACACUUUUGCCAACGGUAAAACGGCUGUGUGCG--- (((.((((((((.----.-------..-))))))).(((.((.(.....).)))))..(((((..((((....))))((((((...))))))...)))))).)))--- ( -26.60) >DroSim_CAF1 26793 91 + 1 UAC-CAGCAAUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCUGC-------ACAGCAACUGUGUGUGUGU (((-((((..(((((...-------)))))..)))).))).((((((...--.........(((.((((....))))..)))-------((((...)))))))))).. ( -25.00) >DroEre_CAF1 1804 91 + 1 UGC-CAGCAGUGUACAAC-------GUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUUUGC-------ACAGCCACUGUGUAUGUGC (((-((((..(((((...-------)))))..)))).))).((((((...--.........(((((((((...((....)).-------....))))))))))))))) ( -30.10) >DroYak_CAF1 25264 98 + 1 UAC-CAGCAAUGUACAACGUACAACGUACAACGCUGCGUAUGCGCAUUAU--UUGAAACGAGCACAGUGGAAACAUUUCUGC-------ACAGCGACUGUGUGUGUGU (((-((((..(((((..........)))))..)))).))).((((((...--.........(((.((((....))))..)))-------((((...)))))))))).. ( -25.70) >DroPer_CAF1 47153 93 + 1 CGCCCAACAAUGU----C-------GU-CAUUGUUACGAAUGCGCAUUACACAUCGAAGCAGCACAGUGGAAACACUUUUGCCAACGGUAAAACGGCUGUGUGCG--- (((.((((((((.----.-------..-))))))).(((.((.(.....).)))))..(((((..((((....))))((((((...))))))...)))))).)))--- ( -26.60) >consensus UAC_CAGCAAUGUACAAC_______GUACAACGCUGCGUAUGCGCAUUAU__UUGAAACGAGCACAGUGGAAACAUUUCUGC_______ACAGCGACUGUGUGUGUGU (((.((((..(((((..........)))))..)))).))).((((((..............(((.((((....))))..)))................)))))).... (-16.40 = -17.59 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:25 2006