
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,867,867 – 10,868,028 |
| Length | 161 |
| Max. P | 0.689476 |

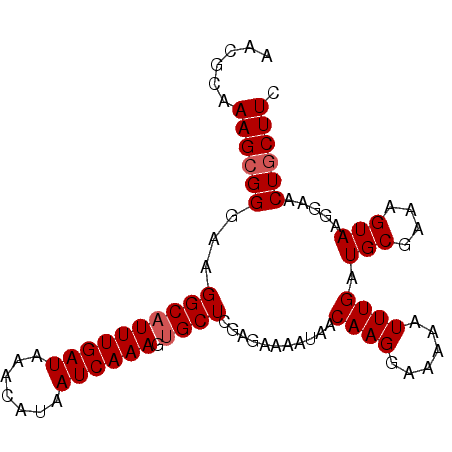
| Location | 10,867,867 – 10,867,958 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -17.45 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10867867 91 - 22224390 AAUGCAAAGCGGGGAGGCAUUUGAUAAACAUAAUCAAAGUGCUCGAGAAAAUAACAAGGAAAAAUUUGAUGCGAAAGUAAGGAACUGCUUC ......((((((.(((.((((((((.......))).))))))))..........((((......)))).(((....))).....)))))). ( -18.20) >DroSec_CAF1 21503 91 - 1 AACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAGUGCUCGAGAAAAUAACAAGAAAAAAUUUGAUGCGAAAGUAAGGAACUGCUUC ......((((((...((((((((((.......))).)))))))..........................(((....))).....)))))). ( -17.50) >DroSim_CAF1 23031 91 - 1 AACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAGUGCUCGAGAAAAUAACAAGGAAAAAUUUGAUGCGAAAGUAAGGAACUGCUUC ......((((((...((((((((((.......))).)))))))...........((((......)))).(((....))).....)))))). ( -17.80) >DroYak_CAF1 21653 91 - 1 AACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAGCGCUCGAGAAAAUAACAAGGAAAAAUUUGAUGCGAAAGUAAGGAACUUCUUC ..(((...)))(((((((.((((((.......))))))))..((..........((((......)))).(((....)))..)).))))).. ( -16.30) >consensus AACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAGUGCUCGAGAAAAUAACAAGGAAAAAUUUGAUGCGAAAGUAAGGAACUGCUUC ......((((((...((((((((((.......)))))).))))...........((((......)))).(((....))).....)))))). (-15.02 = -15.52 + 0.50)


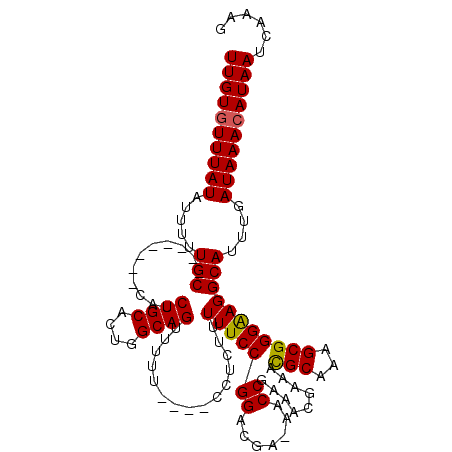
| Location | 10,867,919 – 10,868,028 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10867919 109 - 22224390 UUGUGUUUAUAUUUUUGC--------CAUCUGCACUGGCAGUUUUUUUUUUCUCUUUUUCCGGACGA-AACCGAAACGAAAUGCAAAGCGGGGAGGCAUUUGAUAAACAUAAUCAAAG ((((((((((.....(((--------(.(((((....((((((((..((((.(((......))).))-))..)))))....)))...)))))..))))....))))))))))...... ( -28.00) >DroSec_CAF1 21555 105 - 1 UUGUCUUUAUAUUUUUGC--------CAUCUGCACUGGCAGUUUUU----CUUCUUUUUCCGGACGA-AACCGAAACGAAACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAG .(((((((....((((((--------....(((....)))(((((.----.(((..((((.....))-))..)))..)))))))))))...)))))))((((((.......)))))). ( -22.00) >DroSim_CAF1 23083 105 - 1 UUGUGUUUAUAUUUUUGC--------CAUCUGCACUGGCAGUUUUU----CCUCUUUUUCCGGACGA-AACCGAAACGAAACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAG ((((((((((.....(((--------(..((((....))))...((----((........(((....-..)))........(((...)))))))))))....))))))))))...... ( -28.00) >DroYak_CAF1 21705 117 - 1 UUGUGUUUAUAUUUUUGCCAUCUUGCCAUCUGCACUGGCAGUUUUU-CCUCCUUUUUUUCCGGGCAAAAACCGAAACGAAACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAG ((((((((((.....((((...((((((.......))))))...((-(((.((((.(((((((.......)))....))))...)))).)))))))))....))))))))))...... ( -32.30) >consensus UUGUGUUUAUAUUUUUGC________CAUCUGCACUGGCAGUUUUU____CCUCUUUUUCCGGACGA_AACCGAAACGAAACGCAAAGCGGGAAGGCAUUUGAUAAACAUAAUCAAAG ((((((((((.....(((...........((((....))))...............(((((((.......)).........(((...)))))))))))....))))))))))...... (-20.50 = -20.38 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:21 2006