| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,864,902 – 10,864,997 |
| Length | 95 |
| Max. P | 0.994018 |

| Location | 10,864,902 – 10,864,997 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10864902 95 + 22224390 --AC---CACAGCCCACUGAAACCGGCGAAGAUGAGCAACGUUUGCUUCCAA-----------AGCAGCGGAAACGGAUGCAGAACGGAUGUGG------AAGUGGGCCAGGU-GG--UU --((---(((.(((((((....((.((........)).(((((((.(((...-----------.(((.((....))..))).))))))))))))------.)))))))...))-))--). ( -36.60) >DroSec_CAF1 18616 98 + 1 --ACCACCACAGCCCACUGAAACCGGCGAAGAUGAGCAACGUUUGCUUCCAA-----------AGCAGCGGAAACGGAUGCAGAACGGAUGUGG------AAGUGGGCCAGGU-GG--UU --((((((...(((((((....((.((........)).(((((((.(((...-----------.(((.((....))..))).))))))))))))------.)))))))..)))-))--). ( -39.20) >DroSim_CAF1 20152 97 + 1 --AC-ACGUCAGCCCAUUGAAACCAGAAUGGAUGAUCAAAGUCUGCUUCCAA-----------AGCAGCGGAAACGGAUGCAGAACGGAUGUGG------AAGUGGGCCAGGU-GG--UU --.(-((((((..(((((........))))).))))....(((..(((((..-----------.(((.((....))..)))...((....))))------)))..)))...))-).--.. ( -31.50) >DroYak_CAF1 18758 106 + 1 --ACACACACAGCCCACUGAAAGCGGCGAGGAUGAGCAGAGUUUGCUUCGAA-----------AGCAGCGGAAACGGAUGCAGAACGGAUGUGGAAAUGGAAGUGGGCCAGGU-GGGGUU --...(((...(((((((.....(.((((((.(((((...)))))))))...-----------.(((.((....))..))).........)).).......)))))))...))-)..... ( -32.20) >DroAna_CAF1 16639 104 + 1 UGUC-ACAUCCAACCACU--AACCAGAG-----GGAGAACCUCCACCUCCAGCUACAGCAACCGGAAGCGGAAGCGGAUGCUGAGCGGAAGUGG------CAGCGGGCCAGAAAGG--GU ((((-((.(((.......--.....(((-----(..(.....)..))))..(((.(((((.((....((....)))).))))))))))).))))------))....(((......)--)) ( -37.70) >consensus __AC_ACCACAGCCCACUGAAACCGGCGAAGAUGAGCAACGUUUGCUUCCAA___________AGCAGCGGAAACGGAUGCAGAACGGAUGUGG______AAGUGGGCCAGGU_GG__UU ...........(((((((.......((........)).(((((((.(((...............(((.((....))..))).)))))))))).........)))))))............ (-17.03 = -17.75 + 0.72)

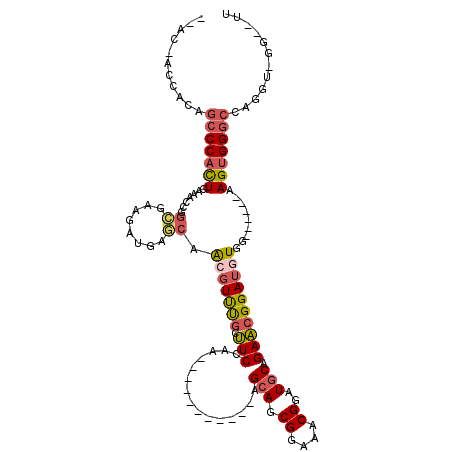
| Location | 10,864,902 – 10,864,997 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.19 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10864902 95 - 22224390 AA--CC-ACCUGGCCCACUU------CCACAUCCGUUCUGCAUCCGUUUCCGCUGCU-----------UUGGAAGCAAACGUUGCUCAUCUUCGCCGGUUUCAGUGGGCUGUG---GU-- .(--((-((..((((((((.------((.....((((.(((.((((...........-----------.)))).)))))))..((........)).))....)))))))))))---))-- ( -31.60) >DroSec_CAF1 18616 98 - 1 AA--CC-ACCUGGCCCACUU------CCACAUCCGUUCUGCAUCCGUUUCCGCUGCU-----------UUGGAAGCAAACGUUGCUCAUCUUCGCCGGUUUCAGUGGGCUGUGGUGGU-- .(--((-((((((((((((.------((.....((((.(((.((((...........-----------.)))).)))))))..((........)).))....))))))))).))))))-- ( -36.20) >DroSim_CAF1 20152 97 - 1 AA--CC-ACCUGGCCCACUU------CCACAUCCGUUCUGCAUCCGUUUCCGCUGCU-----------UUGGAAGCAGACUUUGAUCAUCCAUUCUGGUUUCAAUGGGCUGACGU-GU-- ..--.(-((..((((((...------..........(((((.((((...........-----------.)))).)))))..((((....((.....))..))))))))))...))-).-- ( -25.60) >DroYak_CAF1 18758 106 - 1 AACCCC-ACCUGGCCCACUUCCAUUUCCACAUCCGUUCUGCAUCCGUUUCCGCUGCU-----------UUCGAAGCAAACUCUGCUCAUCCUCGCCGCUUUCAGUGGGCUGUGUGUGU-- .....(-((.(((((((((..............((....(((..((....)).))).-----------..)).((((.....))))................))))))))).)))...-- ( -23.10) >DroAna_CAF1 16639 104 - 1 AC--CCUUUCUGGCCCGCUG------CCACUUCCGCUCAGCAUCCGCUUCCGCUUCCGGUUGCUGUAGCUGGAGGUGGAGGUUCUCC-----CUCUGGUU--AGUGGUUGGAUGU-GACA ..--....((..(((.((((------((((((((((((((((.(((..........))).))))).))).))))))(((((.....)-----)))))).)--))))))..))...-.... ( -42.40) >consensus AA__CC_ACCUGGCCCACUU______CCACAUCCGUUCUGCAUCCGUUUCCGCUGCU___________UUGGAAGCAAACGUUGCUCAUCCUCGCCGGUUUCAGUGGGCUGUGGU_GU__ ...........((((((((...............((...))....(((((((.................)))))))..........................)))))))).......... (-15.35 = -15.19 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:13 2006