
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,862,878 – 10,863,167 |
| Length | 289 |
| Max. P | 0.988031 |

| Location | 10,862,878 – 10,862,984 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10862878 106 + 22224390 CGUGAGCCGAAAGAGACGGGGCAAGUGCUGGCUCUGCUCUGCUGGCCAUUUCGACUUUCGUGUGGCCUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGU .(.((((..(((((((((((((((((....))).)))))))..((((((..((.....)).))))))))))))))))).)...(((((((.......))))))).. ( -40.40) >DroSec_CAF1 16633 92 + 1 CGCGAGCACAAAGAGACGGGGCAAGUGCU--------------GGCCAUUUCGACUUUCGUGUGGCCUUUCUUUGCUCUGCUGGCGCUAACGUGACUUUGGCGCGU ((((....(((((((.(((((((((....--------------((((((..((.....)).))))))....)))))))))))..(((....))).))))).)))). ( -38.20) >DroSim_CAF1 18598 92 + 1 GGCGAGCAGAAAGAGACGGGGCAAGUGCU--------------GGCCAUUUCGACUUUCGUGUGGCCUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGU ((.(((((..((((((...(((....)))--------------((((((..((.....)).))))))))))))))))).))..(((((((.......))))))).. ( -36.60) >DroYak_CAF1 16772 104 + 1 GUCGAGUAGAAAGAGAGAGAGCAGGUGCUGGC--UGCUCUGUUGGCCAUUUCGACAUUCGUGUGGCCUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGU ...(((((((.(((((.(((((((.......)--)))))).))((((((..((.....)).))))))))).))))))).....(((((((.......))))))).. ( -40.10) >consensus CGCGAGCAGAAAGAGACGGGGCAAGUGCU______________GGCCAUUUCGACUUUCGUGUGGCCUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGU ...((((..(((((((...(((....)))..............((((((..((.....)).))))))))))))))))).....(((((((.......))))))).. (-32.58 = -32.20 + -0.38)


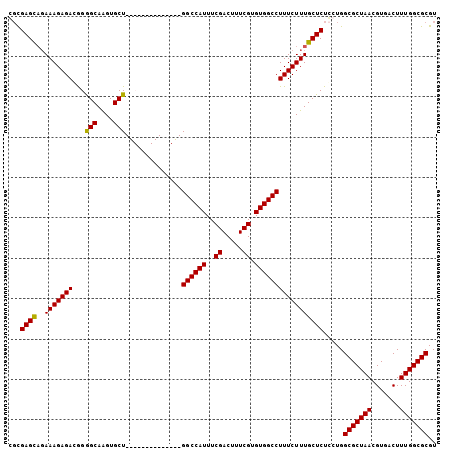
| Location | 10,862,944 – 10,863,058 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
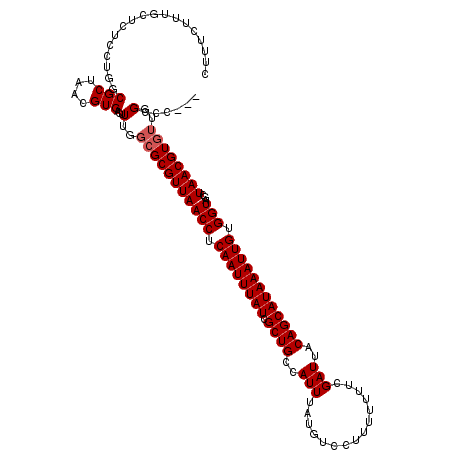
>X_DroMel_CAF1 10862944 114 + 22224390 CUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUA---GCAC--- .......((((....((((..(((((((.(....))))))))(((.((((((((.((((..(((...............)))..)))))))))))).))).))))....)---))).--- ( -28.96) >DroSec_CAF1 16685 117 + 1 CUUUCUUUGCUCUGCUGGCGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC--- .............((..(((((((((((.(....))))))))(((.((((((((.((((..(((...............)))..)))))))))))).))).......))))..))..--- ( -32.06) >DroSim_CAF1 18650 117 + 1 CUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC--- ................(((((....))).))(..(((((((((((.((((((((.((((..(((...............)))..)))))))))))).)))...))))))))..)...--- ( -28.96) >DroYak_CAF1 16836 117 + 1 CUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC--- ................(((((....))).))(..(((((((((((.((((((((.((((..(((...............)))..)))))))))))).)))...))))))))..)...--- ( -28.96) >DroAna_CAF1 13583 110 + 1 CUUUCCUU----------CGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCUCGC .......(----------(((....))))..(..(((((((((((.((((((((.((((..(((...............)))..)))))))))))).)))...))))))))..)...... ( -30.06) >consensus CUUUCUUUGCUCUCCUGGCGCUAACGUGACUUUGGCGCGUUAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC___ .................(((((((.......)))))))....(((.((((((((.((((..(((...............)))..)))))))))))).))).(((((....)))))..... (-24.82 = -25.62 + 0.80)


| Location | 10,862,984 – 10,863,088 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10862984 104 + 22224390 UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUA---GCAC-------ACAC------ACACACUCAUAUAUAUGUGCAUAUAU ..(((.((((((((.((((..(((...............)))..)))))))))))).))).((....)).---((((-------(...------...............)))))...... ( -19.63) >DroSec_CAF1 16725 108 + 1 UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC-------ACAC----ACACACCAUCGCACACACUUGCAUAUA- ..(((.((((((((.((((..(((...............)))..)))))))))))).))).......((((((....-------.)).----))))......(((......))).....- ( -20.96) >DroSim_CAF1 18690 112 + 1 UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC-------ACACACACACACACCAUCGCACACACUUGCAUAUA- ..(((.((((((((.((((..(((...............)))..)))))))))))).))).......((((((....-------.)).))))..........(((......))).....- ( -20.96) >DroYak_CAF1 16876 114 + 1 UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC---GAACACACACACACACACCGUCGCA--CACUUGCAUAUA- ..(((.((((((((.((((..(((...............)))..)))))))))))).))).......(((((.....---.)))))................(((--....))).....- ( -20.96) >DroAna_CAF1 13613 115 + 1 UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCUCGCAAGGAUACACCC--ACACAUCCACAUCCACAUCCACA--- ..(((.((((((((.((((..(((...............)))..)))))))))))).))).......((((.((..((....)).....)).--))))...................--- ( -21.56) >consensus UAACCUCAAUUUAUCGCUGCCAUUUAUGUCCUUUUUUUCGAUUACAGCAUAAAUUGUGGUAGCUAACGUGUUGGCCC_______ACACAC_CACACACCAUCGCACACACUUGCAUAUA_ ..(((.((((((((.((((..(((...............)))..)))))))))))).))).(((((....)))))............................................. (-16.94 = -17.54 + 0.60)



| Location | 10,863,058 – 10,863,167 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10863058 109 + 22224390 ----ACAC------ACACACUCAUAUAUAUGUGCAUAUAUGUAUGUUGGUGUAGCUCACCUGAUAAAUGGUAAACUGCAUGCAUUGUGUAAUCGAUAAAAAAAAAAAAUAAAGUAAAAU ----..((------(((((..((((((((((....)))))))))).))((((((...(((........)))...))))))....))))).............................. ( -21.50) >DroSec_CAF1 16802 103 + 1 ----ACAC----ACACACCAUCGCACACACUUGCAUAUA----UGUUGGUGUAGCUCACCUGAUAAAUGGUAAACUGCAUGCAUUGUGUAAUCGAUAAAA----AAAAUAAAGUAAAAU ----..((----(((...(((.(((.(((((.(((....----))).))))).....(((........)))....))))))...)))))...........----............... ( -18.00) >DroSim_CAF1 18767 106 + 1 ----ACACACACACACACCAUCGCACACACUUGCAUAUA----UGUUGGUGUAGCUCACCUGAUAAAUGGUAAACUGCAUGCAUUGUGUAAUCGAUAAAA-----AAAUAAAGUAAAAU ----......(((((...(((.(((.(((((.(((....----))).))))).....(((........)))....))))))...)))))...........-----.............. ( -18.00) >DroYak_CAF1 16953 104 + 1 GAACACACACACACACACCGUCGCA--CACUUGCAUAUA--------GGUGUAGCUCACCUGAUAAAUGGUAAACUGCAUGCAUUGUGUAAUCGAUAAAA-----AAAUAAAGUAAAAU ...................((((.(--(((.(((((.((--------((((.....)))))).......((.....)))))))..))))...))))....-----.............. ( -18.70) >consensus ____ACAC____ACACACCAUCGCACACACUUGCAUAUA____UGUUGGUGUAGCUCACCUGAUAAAUGGUAAACUGCAUGCAUUGUGUAAUCGAUAAAA_____AAAUAAAGUAAAAU ...................((((...((((.(((((...........((((.....)))).........((.....)))))))..))))...))))....................... (-13.10 = -13.35 + 0.25)


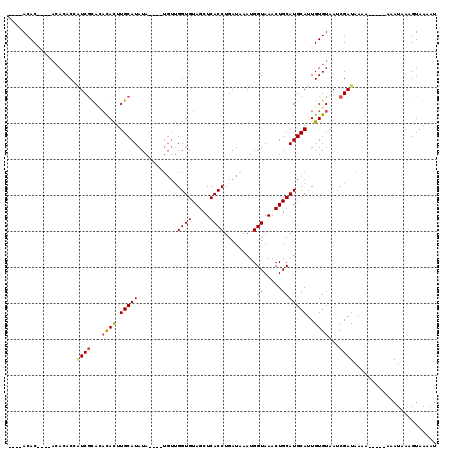
| Location | 10,863,058 – 10,863,167 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.32 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
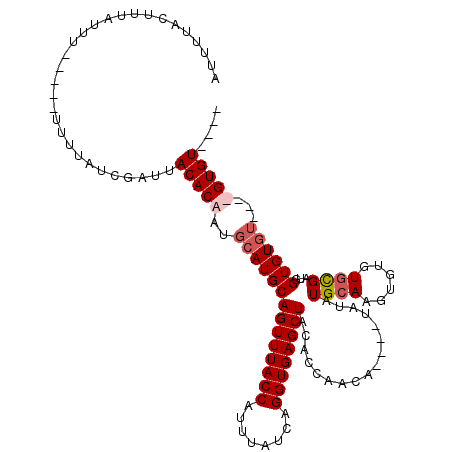
>X_DroMel_CAF1 10863058 109 - 22224390 AUUUUACUUUAUUUUUUUUUUUUAUCGAUUACACAAUGCAUGCAGUUUACCAUUUAUCAGGUGAGCUACACCAACAUACAUAUAUGCACAUAUAUAUGAGUGUGU------GUGU---- .............................(((((......((.((((((((........)))))))).))...((((.((((((((....)))))))).))))))------))).---- ( -23.70) >DroSec_CAF1 16802 103 - 1 AUUUUACUUUAUUUU----UUUUAUCGAUUACACAAUGCAUGCAGUUUACCAUUUAUCAGGUGAGCUACACCAACA----UAUAUGCAAGUGUGUGCGAUGGUGUGU----GUGU---- ...............----..................((((((((((((((........))))))))((((((.((----(((((....)))))))...))))))))----))))---- ( -26.80) >DroSim_CAF1 18767 106 - 1 AUUUUACUUUAUUU-----UUUUAUCGAUUACACAAUGCAUGCAGUUUACCAUUUAUCAGGUGAGCUACACCAACA----UAUAUGCAAGUGUGUGCGAUGGUGUGUGUGUGUGU---- ..............-----...........((((...((((((((((((((........))))))))((((((.((----(((((....)))))))...))))))))))))))))---- ( -29.60) >DroYak_CAF1 16953 104 - 1 AUUUUACUUUAUUU-----UUUUAUCGAUUACACAAUGCAUGCAGUUUACCAUUUAUCAGGUGAGCUACACC--------UAUAUGCAAGUG--UGCGACGGUGUGUGUGUGUGUGUUC ..............-----...........(((((.(((((((((((((((........))))))))(((((--------....((((....--))))..))))))))))))))))).. ( -28.50) >consensus AUUUUACUUUAUUU_____UUUUAUCGAUUACACAAUGCAUGCAGUUUACCAUUUAUCAGGUGAGCUACACCAACA____UAUAUGCAAGUGUGUGCGAUGGUGUGU____GUGU____ ..............................(((((..((((((((((((((........)))))))).................((((......))))...))))))...))))).... (-17.26 = -18.32 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:11 2006