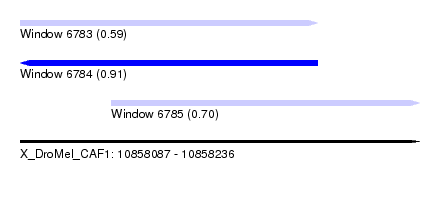
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,858,087 – 10,858,236 |
| Length | 149 |
| Max. P | 0.907673 |

| Location | 10,858,087 – 10,858,198 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.94 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10858087 111 + 22224390 CCUGUCUUUGCCCUUGAAG--CUCGUCAACUUGCUUGUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAAAAA ......(((((..((((((--((.((((....(((((.(((.(.(((((((((((.....))))))))))).).)))))).......)))))).))..)))))).)))))... ( -30.21) >DroSec_CAF1 11903 98 + 1 CCUGUCUUUGCCCUUGAAGCGCUCGCCAACUUGCUUGUCCACGCUGCGUAU---------------ACGUAAUAUGGCAACACCGUCGCUGGCCAGUCCUUCAGAGCAAGAAA ....((((.((..((((((.(((.((((((.((.(((.(((.(.((((...---------------.)))).).))))))))..))...)))).))).)))))).)))))).. ( -26.70) >DroSim_CAF1 13934 113 + 1 CCUGUCUUUGCCCUUGAAGCGCUCGCCGACUUGCUUGUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAGAGA ....(((((((..((((((.(((.((((((......)))...(((((((((((((.....)))))))))))....(....)......)).))).))).)))))).)).))))) ( -38.40) >DroYak_CAF1 12146 111 + 1 CCUGUCUUUGCCCUUGAAG--CUCGCCAACUUGCUUGUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUGCAGCGCAAGUAA .(((.....(((..(((((--...((((....((........))(((((((((((.....)))))))))))...)))).....)))))..)))))).......((....)).. ( -31.50) >consensus CCUGUCUUUGCCCUUGAAG__CUCGCCAACUUGCUUGUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAGAAA ......(((((.((.((((.(((.((((....(((((.(((.(.(((((((((((.....))))))))))).).)))))).......)))))).))).)))))).)))))... (-30.00 = -30.94 + 0.94)

| Location | 10,858,087 – 10,858,198 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.95 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10858087 111 - 22224390 UUUUUUGCUCUGAAGGACUGGCCAGCGAAGGAGUUGCCAUAUUACGUAUACGCACGUUACGUAUACGCAGCGUGGACAAGCAAGUUGACGAG--CUUCAAGGGCAAAGACAGG .((((((((((((((...((((.(((......))))))).....((((((((.......))))))))...(((.(((......))).)))..--)))).)))))))))).... ( -37.40) >DroSec_CAF1 11903 98 - 1 UUUCUUGCUCUGAAGGACUGGCCAGCGACGGUGUUGCCAUAUUACGU---------------AUACGCAGCGUGGACAAGCAAGUUGGCGAGCGCUUCAAGGGCAAAGACAGG ..((((((((((((((.((.((((((.....((((.(((..((.((.---------------...)).))..)))...)))).)))))).))).)))).))))).)))).... ( -35.80) >DroSim_CAF1 13934 113 - 1 UCUCUUGCUCUGAAGGACUGGCCAGCGAAGGAGUUGCCAUAUUACGUAUACGCACGUUACGUAUACGCAGCGUGGACAAGCAAGUCGGCGAGCGCUUCAAGGGCAAAGACAGG (((.((((((((((((.((.(((.((....(..((((((..((.((((((((.......)))))))).))..))).))).)..)).))).))).)))).)))))))))).... ( -40.80) >DroYak_CAF1 12146 111 - 1 UUACUUGCGCUGCAGGACUGGCCAGCGAAGGAGUUGCCAUAUUACGUAUACGCACGUUACGUAUACGCAGCGUGGACAAGCAAGUUGGCGAG--CUUCAAGGGCAAAGACAGG ....((((.((..(((.((.((((((....(..((((((..((.((((((((.......)))))))).))..))).))).)..)))))).))--)))..)).))))....... ( -38.30) >consensus UUUCUUGCUCUGAAGGACUGGCCAGCGAAGGAGUUGCCAUAUUACGUAUACGCACGUUACGUAUACGCAGCGUGGACAAGCAAGUUGGCGAG__CUUCAAGGGCAAAGACAGG ....(((((((((((..((.((((((....(..((((((..((.((((((((.......)))))))).))..))).))).)..)))))).))..)))).)))))))....... (-33.26 = -34.95 + 1.69)



| Location | 10,858,121 – 10,858,236 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10858121 115 + 22224390 GUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAAAAAAAAAAAAAAAAAUGCGACAACAAGAAAAGCGA--AAUGGA (.(((.(.(((((((((((.....))))))))))).).))))...(((((((((((.....))......(((.................)))............)))))--)..))) ( -26.23) >DroSec_CAF1 11939 88 + 1 GUCCACGCUGCGUAU---------------ACGUAAUAUGGCAACACCGUCGCUGGCCAGUCCUUCAGAGCAAGAAAA------------GCGACAACAAGCAGAGCGA--AAUGGA .((((((((((....---------------........((....))..((((((..(..(..(....)..)..)...)------------))))).....))..)))).--..)))) ( -21.40) >DroSim_CAF1 13970 103 + 1 GUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAGAGAA------------GCGACAACAAGCAGAGCGA--AAUGGA (.(((.(.(((((((((((.....))))))))))).).))))...(((((((((.((..(((((((.........)))------------).))).....))..)))))--)..))) ( -33.00) >DroYak_CAF1 12180 105 + 1 GUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUGCAGCGCAAGUAAA------------AGGACAACAAGAAGAGCGUAGAAUGUA .((.(((((((((((((((.....))))))))))....((((..(......)...))))(((((...((....))...------------))))).........))))).))..... ( -36.60) >consensus GUCCACGCUGCGUAUACGUAACGUGCGUAUACGUAAUAUGGCAACUCCUUCGCUGGCCAGUCCUUCAGAGCAAGAAAA____________GCGACAACAAGAAGAGCGA__AAUGGA .....((((((((((((((.....))))))))))....((((..(......)...)))).............................................))))......... (-19.20 = -19.95 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:04 2006