| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,857,553 – 10,857,656 |
| Length | 103 |
| Max. P | 0.601737 |

| Location | 10,857,553 – 10,857,656 |
|---|---|
| Length | 103 |
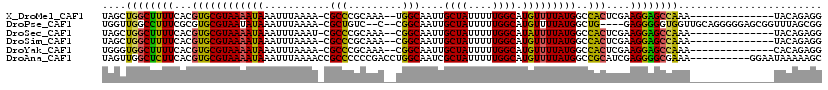
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
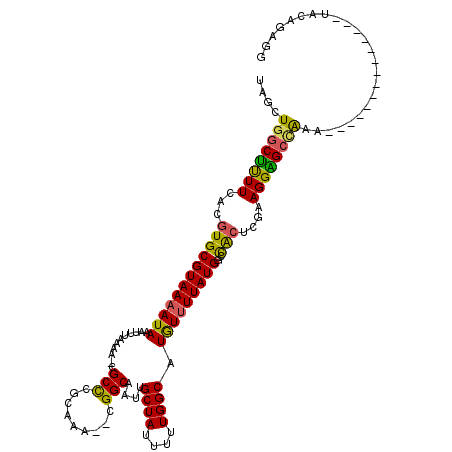
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -21.38 |
| Energy contribution | -20.67 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10857553 103 + 22224390 UAGCUGGCUUUUCACGUGCGUAAAAUAAAUUUAAAA-CGCCCGCAAA--UGGCAAUUGCUAUUUUUGGCAUGUUUUAUGGCCACUCGAAGGAGCCAAA--------------UACAGAGG ....(((((((((..((((((((((((.........-.(((......--.)))...(((((....))))))))))))))..)))..))).))))))..--------------........ ( -25.70) >DroPse_CAF1 32003 111 + 1 UGGUUGGCCUUUCGCGUGCGUAAUAUAAAUUUAAAA-CGCUGUC--C--CGGCAAUUGCUAUUUUUGGCAUGUUUUAUGGCUG----GAGGGGGUGGUUGCAGGGGGAGCGGUUUAGCGG .((....))...(((..((.(((.......)))...-((((.((--(--(.(((((..((.((((..((..........))..----)))).))..))))).)))).))))))...))). ( -38.70) >DroSec_CAF1 11374 103 + 1 UAGCUGGCUUUUCACGUGCGUAAAAUAAAUUUAAAU-CGCCCGCAAA--CGGCAAUUGCUAUUUUUGGCAUAUUUUAUGGCCACUCGAAGGAGCCAAA--------------UACAGAGG ....(((((((((..((((((((((((.........-.(((......--.)))...(((((....))))))))))))))..)))..))).))))))..--------------........ ( -25.80) >DroSim_CAF1 13436 103 + 1 UAGCUGGCUUUUCACGUGCGUAAAAUAAAUUUAAAA-CGCCCGCAAA--CGGCAAUUGCUAUUUUUGGCAUGUUUUAUGGCCACUCGAAGGAGCCAAA--------------UACAGAGG ....(((((((((..((((((((((((.........-.(((......--.)))...(((((....))))))))))))))..)))..))).))))))..--------------........ ( -25.80) >DroYak_CAF1 11620 103 + 1 UGGGUGGCUUUUCACGUGCGUAAAAUAAAUUUAAAA-CGCCCGCAAA--CGGCAAUUGCUAUUUUUGGCAUGUUUUAUGGCCACUCGAAGGAGCCAAA--------------CACAGAGG ((..(((((((((..((((((((((((.........-.(((......--.)))...(((((....))))))))))))))..)))..))).))))))..--------------))...... ( -28.60) >DroAna_CAF1 7480 110 + 1 UAGUUGGCUCUUCACGUGCGUAAAAUAAAUUUAAAACCGCCCCCCGACCUGGCAAUCGCUAUUUUUGGCAUGUUUUAUGGCCGCAUCGAGGGGCGAAA----------GGAAUAAAAAGC ......((.(.....).))..................((((((.(((..((((.((.((((....))))))........))))..))).))))))...----------............ ( -26.20) >consensus UAGCUGGCUUUUCACGUGCGUAAAAUAAAUUUAAAA_CGCCCGCAAA__CGGCAAUUGCUAUUUUUGGCAUGUUUUAUGGCCACUCGAAGGAGCCAAA______________UACAGAGG ....((((((((...((((((((((((...........(((.........)))....((((....)))).)))))))))..)))....))))))))........................ (-21.38 = -20.67 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:01 2006